7Z2J
 
 | | White Bream virus N7-Methyltransferase | | Descriptor: | Non-structural protein 1, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Shannon, A, Gauffre, P, Canard, B, Ferron, F. | | Deposit date: | 2022-02-28 | | Release date: | 2022-11-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.657 Å) | | Cite: | A second type of N7-guanine RNA cap methyltransferase in an unusual locus of a large RNA virus genome.
Nucleic Acids Res., 50, 2022
|
|
2RGN
 
 | | Crystal Structure of p63RhoGEF complex with Galpha-q and RhoA | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, Guanine nucleotide-binding protein G(i) subunit alpha-1,Guanine nucleotide-binding protein G(q) subunit alpha, MAGNESIUM ION, ... | | Authors: | Shankaranarayanan, A, Nance, M.R, Tesmer, J.J.G. | | Deposit date: | 2007-10-04 | | Release date: | 2008-01-15 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structure of Galphaq-p63RhoGEF-RhoA complex reveals a pathway for the activation of RhoA by GPCRs.
Science, 318, 2007
|
|
1YN9
 
 | | Crystal structure of baculovirus RNA 5'-phosphatase complexed with phosphate | | Descriptor: | PHOSPHATE ION, polynucleotide 5'-phosphatase | | Authors: | Changela, A, Martins, A, Shuman, S, Mondragon, A. | | Deposit date: | 2005-01-24 | | Release date: | 2005-02-22 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of baculovirus RNA triphosphatase complexed with phosphate
J.Biol.Chem., 280, 2005
|
|
7KDZ
 
 | |
7KDW
 
 | |
7K6R
 
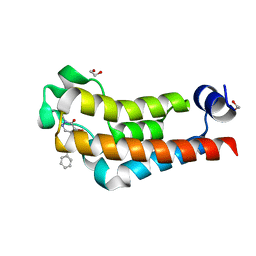 | |
2XEU
 
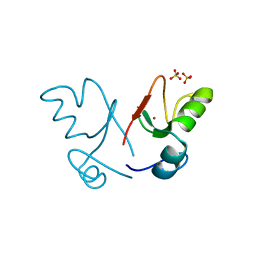 | | Ring domain | | Descriptor: | RING FINGER PROTEIN 4, SULFATE ION, ZINC ION, ... | | Authors: | Plechanovova, A, McMahon, S.A, Johnson, K.A, Navratilova, I, Naismith, J.H, Hay, R.T. | | Deposit date: | 2010-05-18 | | Release date: | 2010-07-28 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Mechanism of Ubiquitylation by Dimeric Ring Ligase Rnf4
Nat.Struct.Mol.Biol., 18, 2011
|
|
7K6S
 
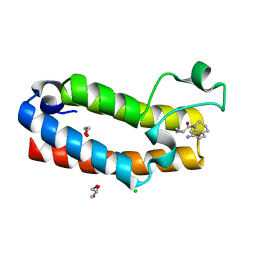 | | Crystal structure of the bromodomain (BD) of human Bromodomain and PHD finger-containing Transcription Factor (BPTF) bound to Compound 4 (SKT1174) | | Descriptor: | (7-amino-3,4-dihydroquinolin-1(2H)-yl)(cyclopropyl)methanone, 1,2-ETHANEDIOL, CHLORIDE ION, ... | | Authors: | Chan, A, Schonbrunn, E. | | Deposit date: | 2020-09-21 | | Release date: | 2020-12-09 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.23 Å) | | Cite: | New inhibitors for the BPTF bromodomain enabled by structural biology and biophysical assay development.
Org.Biomol.Chem., 18, 2020
|
|
2GXB
 
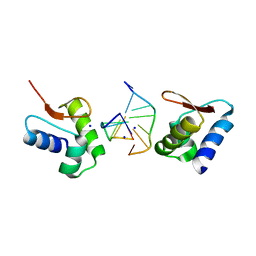 | | Crystal Structure of The Za Domain bound to Z-RNA | | Descriptor: | 5'-R(P*(DU)P*CP*GP*CP*GP*CP*G)-3', Double-stranded RNA-specific adenosine deaminase, SODIUM ION | | Authors: | Athanasiadis, A, Placido, D, Rich, A. | | Deposit date: | 2006-05-08 | | Release date: | 2007-05-01 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | A Left-Handed RNA Double Helix Bound by the Zalpha Domain of the RNA-Editing Enzyme ADAR1.
Structure, 15, 2007
|
|
2O59
 
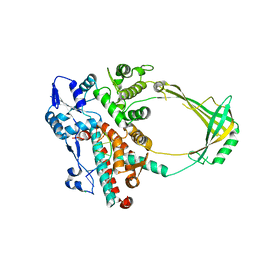 | | Structure of E. coli topoisomerase III in complex with an 8-base single stranded oligonucleotide. Frozen in glycerol pH 8.0 | | Descriptor: | 5'-D(*CP*GP*CP*AP*AP*CP*TP*T)-3', ACETIC ACID, CHLORIDE ION, ... | | Authors: | Changela, A, DiGate, R.J, Mondragon, A. | | Deposit date: | 2006-12-05 | | Release date: | 2007-04-03 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural Studies of E. coli Topoisomerase III-DNA Complexes Reveal a Novel Type IA Topoisomerase-DNA Conformational Intermediate.
J.Mol.Biol., 368, 2007
|
|
2O5C
 
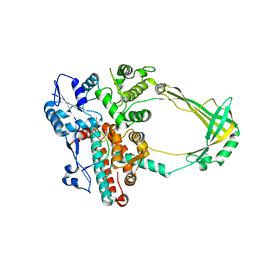 | | Structure of E. coli topoisomerase III in complex with an 8-base single stranded oligonucleotide. Frozen in glucose pH 5.5 | | Descriptor: | 5'-D(*CP*GP*CP*AP*AP*CP*TP*T)-3', CHLORIDE ION, DNA topoisomerase 3, ... | | Authors: | Changela, A, DiGate, R.J, Mondragon, A. | | Deposit date: | 2006-12-05 | | Release date: | 2007-04-03 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural Studies of E. coli Topoisomerase III-DNA Complexes Reveal a Novel Type IA Topoisomerase-DNA Conformational Intermediate.
J.Mol.Biol., 368, 2007
|
|
2O19
 
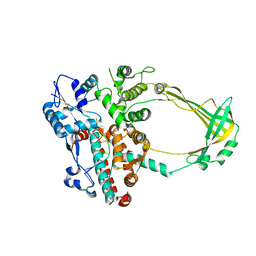 | | Structure of E. coli topoisomersae III in complex with an 8-base single stranded oligonucleotide. Frozen in glycerol at pH 5.5 | | Descriptor: | 5'-D(*CP*GP*CP*AP*AP*CP*TP*T)-3', ACETIC ACID, CHLORIDE ION, ... | | Authors: | Changela, A, DiGate, R, Mondragon, A. | | Deposit date: | 2006-11-28 | | Release date: | 2007-04-03 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structural Studies of E. coli Topoisomerase III-DNA Complexes Reveal a Novel Type IA Topoisomerase-DNA Conformational Intermediate.
J.Mol.Biol., 368, 2007
|
|
3SJF
 
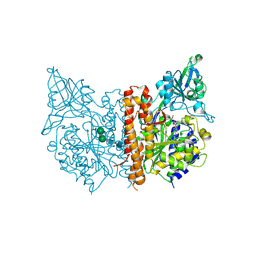 | | X-ray structure of human glutamate carboxypeptidase II in complex with a urea-based inhibitor (A25) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Plechanovova, A, Byun, Y, Alquicer, G, Skultetyova, L, Mlcochova, P, Nemcova, A, Kim, H, Navratil, M, Mease, R, Lubkowski, J, Pomper, M, Konvalinka, J, Rulisek, L, Barinka, C. | | Deposit date: | 2011-06-21 | | Release date: | 2011-10-05 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Novel Substrate-Based Inhibitors of Human Glutamate Carboxypeptidase II with Enhanced Lipophilicity.
J.Med.Chem., 54, 2011
|
|
3SJG
 
 | | Human glutamate carboxypeptidase II (E424A inactive mutant ) in complex with N-acetyl-aspartyl-aminooctanoic acid | | Descriptor: | (2S)-2-[(N-acetyl-L-alpha-aspartyl)amino]nonanoic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Plechanovova, A, Byun, Y, Alquicer, G, Skultetyova, L, Mlcochova, P, Nemcova, A, Kim, H, Navratil, M, Mease, R, Lubkowski, J, Pomper, M, Konvalinka, J, Rulisek, L, Barinka, C. | | Deposit date: | 2011-06-21 | | Release date: | 2011-10-05 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Novel Substrate-Based Inhibitors of Human Glutamate Carboxypeptidase II with Enhanced Lipophilicity.
J.Med.Chem., 54, 2011
|
|
3SJE
 
 | | X-ray structure of human glutamate carboxypeptidase II (the E424A inactive mutant) in complex with N-acetyl-aspartyl-aminononanoic acid | | Descriptor: | (2S)-2-[(N-acetyl-L-alpha-aspartyl)amino]nonanoic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Plechanovova, A, Byun, Y, Alquicer, G, Skultetyova, L, Mlcochova, P, Nemcova, A, Kim, H, Navratil, M, Mease, R, Lubkowski, J, Pomper, M, Konvalinka, J, Rulisek, L, Barinka, C. | | Deposit date: | 2011-06-21 | | Release date: | 2011-10-05 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Novel Substrate-Based Inhibitors of Human Glutamate Carboxypeptidase II with Enhanced Lipophilicity.
J.Med.Chem., 54, 2011
|
|
4AP4
 
 | | Rnf4 - ubch5a - ubiquitin heterotrimeric complex | | Descriptor: | E3 UBIQUITIN LIGASE RNF4, UBIQUITIN C, UBIQUITIN-CONJUGATING ENZYME E2 D1, ... | | Authors: | Plechanovova, A, Hay, R.T, Tatham, M.H, Jaffray, E, Naismith, J.H. | | Deposit date: | 2012-03-30 | | Release date: | 2012-07-25 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Structure of a Ring E3 Ligase and Ubiquitin-Loaded E2 Primed for Catalysis
Nature, 489, 2012
|
|
3SJX
 
 | | X-ray structure of human glutamate carboxypeptidase II (the E424A inactive mutant) in complex with N-acetyl-aspartyl-methionine | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Plechanovova, A, Byun, Y, Alquicer, G, Skultetyova, L, Mlcochova, P, Nemcova, A, Kim, H, Navratil, M, Mease, R, Lubkowski, J, Pomper, M, Konvalinka, J, Rulisek, L, Barinka, C. | | Deposit date: | 2011-06-22 | | Release date: | 2011-10-05 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Novel Substrate-Based Inhibitors of Human Glutamate Carboxypeptidase II with Enhanced Lipophilicity.
J.Med.Chem., 54, 2011
|
|
6IF7
 
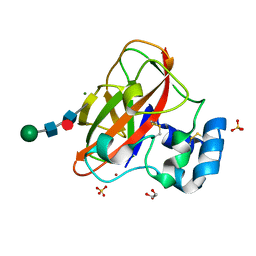 | | Crystal Structure of AA10 Lytic Polysaccharide Monooxygenase from Tectaria macrodonta | | Descriptor: | COPPER (II) ION, Chitin binding protein, GLYCEROL, ... | | Authors: | Archana, A, Yadav, S.K, Singh, P.K, Vasudev, P.G. | | Deposit date: | 2018-09-18 | | Release date: | 2019-04-24 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Insecticidal fern protein Tma12 is possibly a lytic polysaccharide monooxygenase.
Planta, 249, 2019
|
|
2N0R
 
 | |
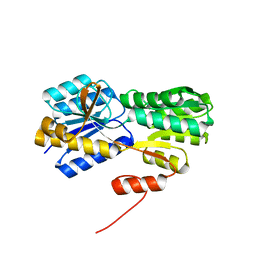
2H3H
 
 | |
4JMQ
 
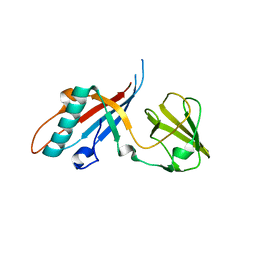 | | Crystal structure of pb9: The Dit of bacteriophage T5. | | Descriptor: | Bacteriophage T5 distal tail protein | | Authors: | Flayhan, A, Vellieux, F.M.D, Girard, E, Maury, O, Boulanger, P, Breyton, C. | | Deposit date: | 2013-03-14 | | Release date: | 2013-11-06 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.895 Å) | | Cite: | Crystal Structure of pb9, the Distal Tail Protein of Bacteriophage T5: a Conserved Structural Motif among All Siphophages.
J.Virol., 88, 2014
|
|
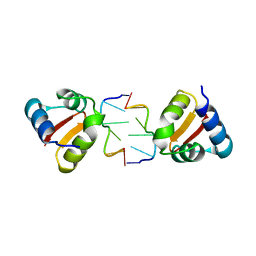
4KA4
 
 | |
7NYQ
 
 | | Crystal structure of the Mei-P26 NHL domain | | Descriptor: | Meiotic P26, isoform C | | Authors: | Salerno-Kochan, A, Gaik, M, Medenbach, J, Glatt, S. | | Deposit date: | 2021-03-23 | | Release date: | 2022-05-04 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Molecular insights into RNA recognition and gene regulation by the TRIM-NHL protein Mei-P26.
Life Sci Alliance, 5, 2022
|
|
7O2L
 
 | | Yeast 20S proteasome in complex with the covalently bound inhibitor b-lactone (2R,3S)-3-isopropyl-4-oxo-2-oxetane-carboxylate (IOC) | | Descriptor: | (2 {R},3 {S})-3-methanoyl-4-methyl-2-hydroxy-pentanoic acid, 20S proteasome, BJ4_G0020160.mRNA.1.CDS.1, ... | | Authors: | Shi, Y.M, Hirschmann, M, Shi, Y.N, Shabbir, A, Abebew, D, Tobias, N.J, Gruen, P, Crames, J.J, Poeschel, L, Kuttenlochner, W, Richter, C, Herrmann, J, Mueller, R, Thanwisai, A, Pidot, S.J, Stinear, T.P, Groll, M, Kim, Y, Bode, H. | | Deposit date: | 2021-03-30 | | Release date: | 2022-04-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Global analysis of biosynthetic gene clusters reveals conserved and unique natural products in entomopathogenic nematode-symbiotic bacteria.
Nat.Chem., 14, 2022
|
|
2VUX
 
 | | Human ribonucleotide reductase, subunit M2 B | | Descriptor: | FE (III) ION, RIBONUCLEOSIDE-DIPHOSPHATE REDUCTASE SUBUNIT M2 B | | Authors: | Welin, M, Moche, M, Andersson, J, Arrowsmith, C.H, Berglund, H, Busam, R.D, Collins, R, Dahlgren, L.G, Edwards, A.M, Flodin, S, Flores, A, Graslund, S, Hammarstrom, M, Herman, M.D, Johansson, A, Johansson, I, Kallas, A, Karlberg, T, Kotenyova, T, Lehtio, L, Nilsson, M.E, Nyman, T, Persson, C, Sagemark, J, Schueler, H, Svensson, L, Thorsell, A.G, Tresaugues, L, van Den Berg, S, Weigelt, J, Wikstrom, M, Nordlund, P, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-05-31 | | Release date: | 2008-07-15 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Human Ribonucleotide Reductase, Subunit M2 B
To be Published
|
|
