4ALI
 
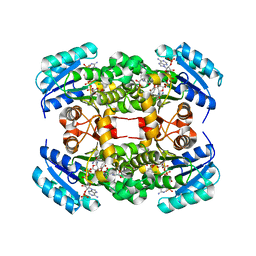 | | Crystal structure of S. aureus FabI in complex with NADP and triclosan (P1) | | Descriptor: | ENOYL-[ACYL-CARRIER-PROTEIN] REDUCTASE [NADPH], GLUTAMIC ACID, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Schiebel, J, Chang, A, Tonge, P.J, Kisker, C. | | Deposit date: | 2012-03-04 | | Release date: | 2012-05-09 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Staphylococcus Aureus Fabi: Inhibition, Substrate Recognition and Potential Implications for in Vivo Essentiality
Structure, 20, 2012
|
|
4ALK
 
 | | Crystal structure of S. aureus FabI in complex with NADP and 5-ethyl- 2-phenoxyphenol | | Descriptor: | 5-ETHYL-2-PHENOXYPHENOL, ENOYL-[ACYL-CARRIER-PROTEIN] REDUCTASE [NADPH], GLUTAMIC ACID, ... | | Authors: | Schiebel, J, Chang, A, Tonge, P.J, Kisker, C. | | Deposit date: | 2012-03-04 | | Release date: | 2012-05-09 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Staphylococcus Aureus Fabi: Inhibition, Substrate Recognition and Potential Implications for in Vivo Essentiality
Structure, 20, 2012
|
|
4ALJ
 
 | | Crystal structure of S. aureus FabI in complex with NADP and 5-chloro- 2-phenoxyphenol | | Descriptor: | 5-CHLORO-2-PHENOXYPHENOL, ENOYL-[ACYL-CARRIER-PROTEIN] REDUCTASE [NADPH], GLUTAMIC ACID, ... | | Authors: | Schiebel, J, Chang, A, Tonge, P.J, Kisker, C. | | Deposit date: | 2012-03-04 | | Release date: | 2012-05-09 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Staphylococcus Aureus Fabi: Inhibition, Substrate Recognition and Potential Implications for in Vivo Essentiality
Structure, 20, 2012
|
|
4ALL
 
 | | Crystal structure of S. aureus FabI in complex with NADP and triclosan (P212121) | | Descriptor: | ENOYL-[ACYL-CARRIER-PROTEIN] REDUCTASE [NADPH], NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, TRICLOSAN | | Authors: | Schiebel, J, Chang, A, Tonge, P.J, Kisker, C. | | Deposit date: | 2012-03-04 | | Release date: | 2012-05-09 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Staphylococcus Aureus Fabi: Inhibition, Substrate Recognition and Potential Implications for in Vivo Essentiality
Structure, 20, 2012
|
|
2OGM
 
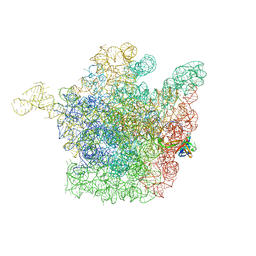 | | The crystal structure of the large ribosomal subunit from Deinococcus radiodurans complexed with the pleuromutilin derivative SB-571519 | | Descriptor: | (2S,3AR,4R,5S,6S,8R,9R,9AR,10R)-2,5-DIHYDROXY-4,6,9,10-TETRAMETHYL-1-OXO-6-VINYLDECAHYDRO-3A,9-PROP[1]ENOCYCLOPENTA[8]ANNULEN-8-YL [(6-AMINOPYRIDAZIN-3-YL)CARBONYL]CARBAMATE, 23S ribosomal RNA, 50S ribosomal protein L3 | | Authors: | Davidovich, C, Bashan, A, Auerbach-Nevo, T, Yonath, A. | | Deposit date: | 2007-01-07 | | Release date: | 2007-05-01 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Induced-fit tightens pleuromutilins binding to ribosomes and remote interactions enable their selectivity.
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
2OGN
 
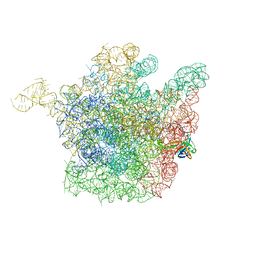 | | The crystal structure of the large ribosomal subunit from Deinococcus radiodurans complexed with the pleuromutilin derivative SB-280080 | | Descriptor: | (3AS,4R,5S,6S,8R,9R,9AR,10R)-5-HYDROXY-4,6,9,10-TETRAMETHYL-1-OXO-6-VINYLDECAHYDRO-3A,9-PROPANOCYCLOPENTA[8]ANNULEN-8-YL (PIPERIDIN-4-YLTHIO)ACETATE, 23S ribosomal RNA, 50S ribosomal protein L3 | | Authors: | Davidovich, C, Bashan, A, Auerbach-Nevo, T, Yonath, A. | | Deposit date: | 2007-01-07 | | Release date: | 2007-05-01 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3.56 Å) | | Cite: | Induced-fit tightens pleuromutilins binding to ribosomes and remote interactions enable their selectivity.
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
2OGO
 
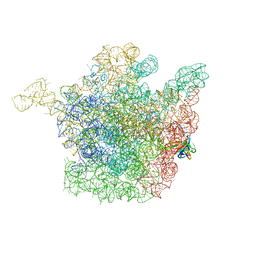 | | The crystal structure of the large ribosomal subunit from Deinococcus radiodurans complexed with the pleuromutilin derivative retapamulin (SB-275833) | | Descriptor: | 23S ribosomal RNA, 50S ribosomal protein L3, Retapamulin | | Authors: | Davidovich, C, Bashan, A, Auerbach-Nevo, T, Yonath, A. | | Deposit date: | 2007-01-07 | | Release date: | 2007-05-01 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3.66 Å) | | Cite: | Induced-fit tightens pleuromutilins binding to ribosomes and remote interactions enable their selectivity.
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
4D44
 
 | | Crystal structure of S. aureus FabI in complex with NADP and 5-ethyl- 4-fluoro-2-((2-fluoropyridin-3-yl)oxy)phenol | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, 5-ethyl-4-fluoro-2-[(2-fluoropyridin-3-yl)oxy]phenol, ENOYL-[ACYL-CARRIER-PROTEIN] REDUCTASE [NADPH], ... | | Authors: | Schiebel, J, Chang, A, Tonge, P.J, Sotriffer, C.A, Kisker, C. | | Deposit date: | 2014-10-26 | | Release date: | 2015-03-04 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | An Ordered Water Channel in Staphylococcus Aureus Fabi: Unraveling the Mechanism of Substrate Recognition and Reduction.
Biochemistry, 54, 2015
|
|
4CV2
 
 | | Crystal structure of E. coli FabI in complex with NADH and CG400549 | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, 1-(3-amino-2-methylbenzyl)-4-[2-(thiophen-2-yl)ethoxy]pyridin-2(1H)-one, ENOYL-[ACYL-CARRIER-PROTEIN] REDUCTASE [NADH] | | Authors: | Tareilus, M, Schiebel, J, Chang, A, Shah, S, Tonge, P.J, Kisker, C. | | Deposit date: | 2014-03-22 | | Release date: | 2014-04-16 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Rational Design of Broad Spectrum Antibacterial Activity Based on a Clinically Relevant Enoyl-Acyl Carrier Protein (Acp) Reductase Inhibitor.
J.Biol.Chem., 289, 2014
|
|
4CV1
 
 | | Crystal structure of S. aureus FabI in complex with NADPH and CG400549 | | Descriptor: | 1,2-ETHANEDIOL, 1-(3-amino-2-methylbenzyl)-4-[2-(thiophen-2-yl)ethoxy]pyridin-2(1H)-one, CHLORIDE ION, ... | | Authors: | Schiebel, J, Chang, A, Shah, S, Tonge, P.J, Kisker, C. | | Deposit date: | 2014-03-22 | | Release date: | 2014-04-16 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Rational Design of Broad Spectrum Antibacterial Activity Based on a Clinically Relevant Enoyl-Acyl Carrier Protein (Acp) Reductase Inhibitor.
J.Biol.Chem., 289, 2014
|
|
4D45
 
 | | Crystal structure of S. aureus FabI in complex with NADP and 5-bromo- 2-(4-chloro-2-hydroxyphenoxy)benzonitrile | | Descriptor: | 5-bromo-2-(4-chloro-2-hydroxyphenoxy)benzonitrile, ENOYL-[ACYL-CARRIER-PROTEIN] REDUCTASE [NADPH], GLUTAMIC ACID, ... | | Authors: | Tareilus, M, Schiebel, J, Chang, A, Tonge, P.J, Sotriffer, C.A, Kisker, C. | | Deposit date: | 2014-10-26 | | Release date: | 2015-03-04 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | An Ordered Water Channel in Staphylococcus Aureus Fabi: Unraveling the Mechanism of Substrate Recognition and Reduction.
Biochemistry, 54, 2015
|
|
4CV0
 
 | | Crystal structure of S. aureus FabI in complex with NADPH and CG400549 (small unit cell) | | Descriptor: | 1,2-ETHANEDIOL, 1-(3-amino-2-methylbenzyl)-4-[2-(thiophen-2-yl)ethoxy]pyridin-2(1H)-one, ENOYL-[ACYL-CARRIER-PROTEIN] REDUCTASE [NADPH], ... | | Authors: | Schiebel, J, Chang, A, Shah, S, Tonge, P.J, Kisker, C. | | Deposit date: | 2014-03-22 | | Release date: | 2014-04-16 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Rational Design of Broad Spectrum Antibacterial Activity Based on a Clinically Relevant Enoyl-Acyl Carrier Protein (Acp) Reductase Inhibitor.
J.Biol.Chem., 289, 2014
|
|
4CV3
 
 | | Crystal structure of E. coli FabI in complex with NADH and PT166 | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, 2-hexyl-1-methyl-5-(2-methylphenoxy)pyridin-4(1H)-one, ENOYL-[ACYL-CARRIER-PROTEIN] REDUCTASE [NADH] | | Authors: | Eltschkner, S, Schiebel, J, Chang, A, Shah, S, Tonge, P.J, Kisker, C. | | Deposit date: | 2014-03-22 | | Release date: | 2014-04-16 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Rational Design of Broad Spectrum Antibacterial Activity Based on a Clinically Relevant Enoyl-Acyl Carrier Protein (Acp) Reductase Inhibitor.
J.Biol.Chem., 289, 2014
|
|
4D41
 
 | | Crystal structure of S. aureus FabI in complex with NADP and 5-hexyl- 2-(4-nitrophenoxy)phenol | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, 5-HEXYL-2-(4-NITROPHENOXY)PHENOL, ... | | Authors: | Schiebel, J, Chang, A, Tonge, P.J, Sotriffer, C.A, Kisker, C. | | Deposit date: | 2014-10-26 | | Release date: | 2015-03-04 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | An Ordered Water Channel in Staphylococcus Aureus Fabi: Unraveling the Mechanism of Substrate Recognition and Reduction.
Biochemistry, 54, 2015
|
|
4D46
 
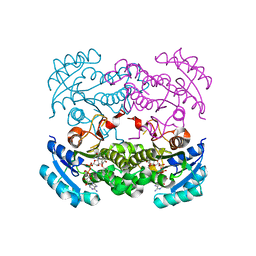 | | Crystal structure of E. coli FabI in complex with NAD and 5-bromo-2-(4-chloro-2-hydroxyphenoxy)benzonitrile | | Descriptor: | 5-bromo-2-(4-chloro-2-hydroxyphenoxy)benzonitrile, ENOYL-[ACYL-CARRIER-PROTEIN] REDUCTASE [NADH], NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Tareilus, M, Schiebel, J, Chang, A, Tonge, P.J, Sotriffer, C.A, Kisker, C. | | Deposit date: | 2014-10-26 | | Release date: | 2015-03-04 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | An Ordered Water Channel in Staphylococcus Aureus Fabi: Unraveling the Mechanism of Substrate Recognition and Reduction.
Biochemistry, 54, 2015
|
|
4D42
 
 | | Crystal structure of S. aureus FabI in complex with NADP and 4-fluoro- 5-hexyl-2-phenoxyphenol | | Descriptor: | 4-fluoro-5-hexyl-2-phenoxyphenol, ENOYL-[ACYL-CARRIER-PROTEIN] REDUCTASE [NADPH], GLUTAMIC ACID, ... | | Authors: | Schiebel, J, Chang, A, Tonge, P.J, Sotriffer, C.A, Kisker, C. | | Deposit date: | 2014-10-26 | | Release date: | 2015-03-04 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | An ordered water channel in Staphylococcus aureus FabI: unraveling the mechanism of substrate recognition and reduction.
Biochemistry, 54, 2015
|
|
4CUZ
 
 | | Crystal structure of S. aureus FabI in complex with NADPH and PT173 | | Descriptor: | 1-(3-amino-2-methylbenzyl)-4-hexylpyridin-2(1H)-one, ENOYL-ACP REDUCTASE MOLECULE ENOYL-[ACYL-CARRIER-PROTEIN] REDUCTASE [NADPH], NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Schiebel, J, Chang, A, Shah, S, Tonge, P.J, Kisker, C. | | Deposit date: | 2014-03-22 | | Release date: | 2014-04-16 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Rational Design of Broad Spectrum Antibacterial Activity Based on a Clinically Relevant Enoyl-Acyl Carrier Protein (Acp) Reductase Inhibitor.
J.Biol.Chem., 289, 2014
|
|
2CIJ
 
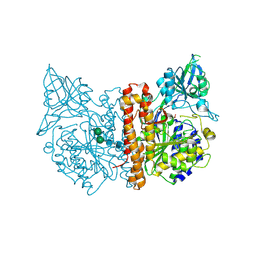 | | membrane-bound glutamate carboxypeptidase II (GCPII) with bound methionine | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Barinka, C, Plechanovova, A, Rulisek, L, Mlcochova, P, Majer, P, Slusher, B.S, Hilgenfeld, R, Mesters, J.R, Konvalinka, J. | | Deposit date: | 2006-03-21 | | Release date: | 2007-03-27 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Glutamate Carboxypeptidase II
Handbook of Metalloproteins, 4, 2011
|
|
4D43
 
 | | Crystal structure of S. aureus FabI in complex with NADP and 2-(2- chloro-4-nitrophenoxy)-5-ethyl-4-fluorophenol | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, 2-(2-chloro-4-nitrophenoxy)-5-ethyl-4-fluorophenol, ENOYL-[ACYL-CARRIER-PROTEIN] REDUCTASE [NADPH], ... | | Authors: | Schiebel, J, Chang, A, Tonge, P.J, Sotriffer, C.A, Kisker, C. | | Deposit date: | 2014-10-26 | | Release date: | 2015-03-04 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | An Ordered Water Channel in Staphylococcus Aureus Fabi: Unraveling the Mechanism of Substrate Recognition and Reduction.
Biochemistry, 54, 2015
|
|
2NCS
 
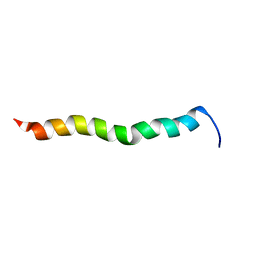 | | NMR assignment and structure of a peptide derived from the membrane proximal external region of HIV-1 gp41 in the presence of dodecylphosphocholine micelles | | Descriptor: | Envelope glycoprotein gp41 | | Authors: | Jimenez, M, Nieva, J.L, Rujas, E, Partida-Hanon, A, Bruix, M. | | Deposit date: | 2016-04-14 | | Release date: | 2017-02-22 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural basis for broad neutralization of HIV-1 through the molecular recognition of 10E8 helical epitope at the membrane interface.
Sci Rep, 6, 2016
|
|
4OEI
 
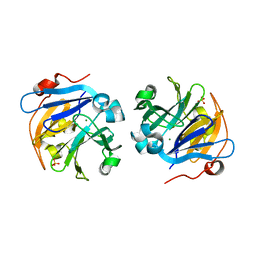 | | Crystal structure of plant lectin from Cicer arietinum at 2.6 angstrom resolution | | Descriptor: | Lectin, MAGNESIUM ION, SULFATE ION | | Authors: | Kumar, S, Dube, D, Bhushan, A, Dey, S, Sharma, S, Singh, T.P. | | Deposit date: | 2014-01-13 | | Release date: | 2014-02-05 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of plant lectin from Cicer arietinum at
2.6 angstrom resolution
TO BE PUBLISHED
|
|
4ORV
 
 | | Crystal structure of the ternary complex of camel peptidoglycan recognition protein PGRP-S with 7- phenylheptanoic acid and N- acetylglucosamine at 2.50 A resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 7-phenylheptanoic acid, GLYCEROL, ... | | Authors: | Yamini, S, Sharma, P, Yadav, S.P, Sinha, M, Bhushan, A, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2014-02-12 | | Release date: | 2014-03-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of the ternary complex of camel peptidoglycan recognition protein PGRP-S with 7- phenylheptanoic acid and N- acetylglucosamine at 2.50 A resolution
To be Published
|
|
4NJB
 
 | | Crystal structure of the complex of lactoperoxidase from bovine with 3,3-oxydipyridine at 2.31 A resolution | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Yamini, S, Sirohi, H.V, Sinha, M, Bhushan, A, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2013-11-09 | | Release date: | 2013-11-27 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Crystal structure of the complex of lactoperoxidase from bovine with 3,3-oxydipyridine at 2.31 A resolution
To be Published
|
|
2DSU
 
 | | Binding of chitin-like polysaccharide to protective signalling factor: Crystal structure of the complex formed between signalling protein from sheep (SPS-40) with a tetrasaccharide at 2.2 A resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-alpha-D-glucopyranose, Chitinase-3-like protein 1, alpha-D-mannopyranose-(1-4)-beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-alpha-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Srivastava, D.B, Ethayathulla, A.S, Kumar, J, Singh, N, Sharma, S, Kaur, P, Bhushan, A, Singh, T.P. | | Deposit date: | 2006-07-07 | | Release date: | 2006-07-25 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Carbohydrate binding properties and carbohydrate induced conformational switch in sheep secretory glycoprotein (SPS-40): crystal structures of four complexes of SPS-40 with chitin-like oligosaccharides
J.Struct.Biol., 158, 2007
|
|
2M9U
 
 | | Solution NMR structure of the C-terminal domain (CTD) of Moloney murine leukemia virus integrase, Northeast Structural Genomics Target OR41A | | Descriptor: | Integrase p46 | | Authors: | Aiyer, S, Rossi, P, Schneider, W.M, Chander, A, Roth, M.J, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2013-06-19 | | Release date: | 2013-12-18 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Altering murine leukemia virus integration through disruption of the integrase and BET protein family interaction.
Nucleic Acids Res., 42, 2014
|
|
