7MCL
 
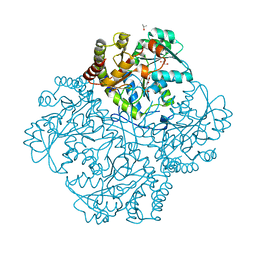 | | Crystal structure of Staphylococcus aureus Cystathionine gamma-lyase, PLP bound | | Descriptor: | Bifunctional cystathionine gamma-lyase/homocysteine desulfhydrase, GLYCEROL, PYRIDOXAL-5'-PHOSPHATE, ... | | Authors: | Nuthanakanti, A, Serganov, A, Kaushik, A. | | Deposit date: | 2021-04-02 | | Release date: | 2021-06-23 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Inhibitors of bacterial H 2 S biogenesis targeting antibiotic resistance and tolerance.
Science, 372, 2021
|
|
7MCY
 
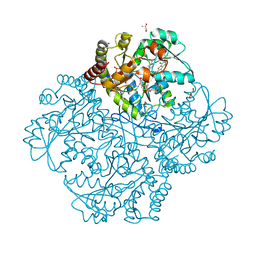 | | Crystal structure of Staphylococcus aureus Cystathionine gamma lyase, Holoenzyme with bound NL3 | | Descriptor: | 3-{[6-(7-chloro-1-benzothiophen-2-yl)-1H-indol-1-yl]methyl}-1H-pyrazole-5-carboxylic acid, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Bifunctional cystathionine gamma-lyase/homocysteine desulfhydrase, ... | | Authors: | Nuthanakanti, A, Serganov, A, Kaushik, A. | | Deposit date: | 2021-04-02 | | Release date: | 2021-06-23 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Inhibitors of bacterial H 2 S biogenesis targeting antibiotic resistance and tolerance.
Science, 372, 2021
|
|
7MCU
 
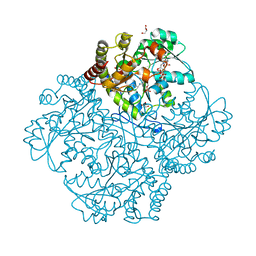 | | Crystal structure of Staphylococcus aureus Cystathionine gamma lyase, Holoenzyme with bound NL2 | | Descriptor: | 5-[(6-bromo-1H-indol-1-yl)methyl]-2-methylfuran-3-carboxylic acid, Bifunctional cystathionine gamma-lyase/homocysteine desulfhydrase, CITRATE ANION, ... | | Authors: | Nuthanakanti, A, Serganov, A, Kaushik, A. | | Deposit date: | 2021-04-02 | | Release date: | 2021-06-23 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Inhibitors of bacterial H 2 S biogenesis targeting antibiotic resistance and tolerance.
Science, 372, 2021
|
|
7MD8
 
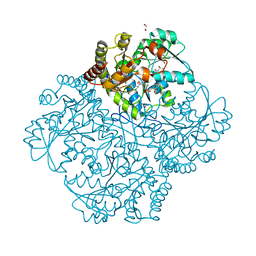 | | Crystal structure of Staphylococcus aureus cystathionine gamma lyase holoenzyme Y103N mutant co-crystallized with NL2 | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Bifunctional cystathionine gamma-lyase/homocysteine desulfhydrase, GLYCEROL, ... | | Authors: | Nuthanakanti, A, Serganov, A, Kaushik, A. | | Deposit date: | 2021-04-03 | | Release date: | 2021-06-23 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Inhibitors of bacterial H 2 S biogenesis targeting antibiotic resistance and tolerance.
Science, 372, 2021
|
|
7MDA
 
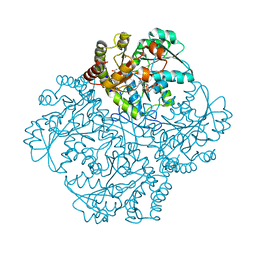 | | Crystal structure of Staphylococcus aureus cystathionine gamma lyase holoenzyme Y103A mutant co-crystallized with NL1 | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Bifunctional cystathionine gamma-lyase/homocysteine desulfhydrase, N-[(6-bromo-1H-indol-1-yl)acetyl]glycine, ... | | Authors: | Nuthanakanti, A, Serganov, A, Kaushik, A. | | Deposit date: | 2021-04-03 | | Release date: | 2021-06-23 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Inhibitors of bacterial H 2 S biogenesis targeting antibiotic resistance and tolerance.
Science, 372, 2021
|
|
7ED5
 
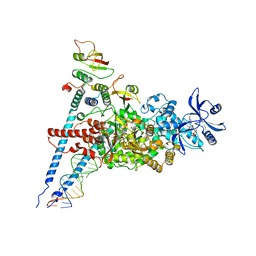 | | A dual mechanism of action of AT-527 against SARS-CoV-2 polymerase | | Descriptor: | MAGNESIUM ION, Non-structural protein 7, Non-structural protein 8, ... | | Authors: | Shannon, A, Fattorini, V, Sama, B, Selisko, B, Feracci, M, Falcou, C, Gauffre, P, El Kazzi, P, Delpal, A, Decroly, E, Alvarez, K, Eydoux, C, Guillemot, J.-C, Moussa, A, Good, S, Colla, P, Lin, K, Sommadossi, J.-P, Zhu, Y.X, Yan, X.D, Shi, H, Ferron, F, Canard, B. | | Deposit date: | 2021-03-15 | | Release date: | 2022-02-16 | | Last modified: | 2024-04-10 | | Method: | ELECTRON MICROSCOPY (2.98 Å) | | Cite: | A dual mechanism of action of AT-527 against SARS-CoV-2 polymerase.
Nat Commun, 13, 2022
|
|
8G9W
 
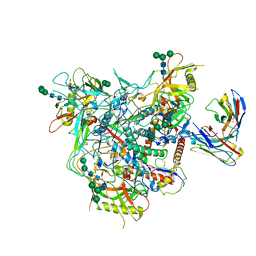 | | Cryo-EM structure of vFP49.02 Fab in complex with HIV-1 Env BG505 DS-SOSIP.664 (conformation 1) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope glycoprotein gp120, ... | | Authors: | Changela, A, Gorman, J, Kwong, P.D. | | Deposit date: | 2023-02-22 | | Release date: | 2023-04-19 | | Last modified: | 2023-06-14 | | Method: | ELECTRON MICROSCOPY (4.66 Å) | | Cite: | Diverse Murine Vaccinations Reveal Distinct Antibody Classes to Target Fusion Peptide and Variation in Peptide Length to Improve HIV Neutralization.
J.Virol., 97, 2023
|
|
8G9Y
 
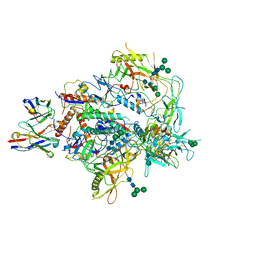 | | Cryo-EM structure of vFP49.02 Fab in complex with HIV-1 Env BG505 DS-SOSIP.664 (conformation 3) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope glycoprotein gp120, ... | | Authors: | Changela, A, Gorman, J, Kwong, P.D. | | Deposit date: | 2023-02-22 | | Release date: | 2023-04-19 | | Last modified: | 2023-06-14 | | Method: | ELECTRON MICROSCOPY (4.28 Å) | | Cite: | Diverse Murine Vaccinations Reveal Distinct Antibody Classes to Target Fusion Peptide and Variation in Peptide Length to Improve HIV Neutralization.
J.Virol., 97, 2023
|
|
8G9X
 
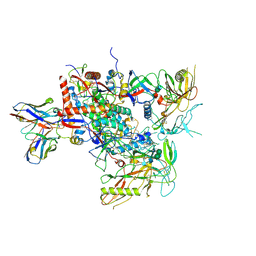 | | Cryo-EM structure of vFP49.02 Fab in complex with HIV-1 Env BG505 DS-SOSIP.664 (conformation 2) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope glycoprotein gp120, ... | | Authors: | Changela, A, Gorman, J, Kwong, P.D. | | Deposit date: | 2023-02-22 | | Release date: | 2023-04-19 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (4.46 Å) | | Cite: | Diverse Murine Vaccinations Reveal Distinct Antibody Classes to Target Fusion Peptide and Variation in Peptide Length to Improve HIV Neutralization.
J.Virol., 97, 2023
|
|
8U9L
 
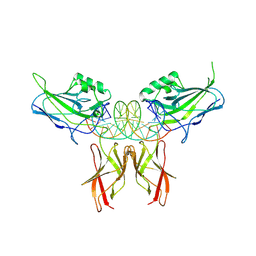 | | Crystal Structure of RelA-cRel chimera complex with DNA | | Descriptor: | DNA (5'-D(P*GP*AP*AP*TP*CP*GP*GP*AP*AP*AP*TP*TP*CP*CP*CP*AP*TP*CP*AP*A)-3'), DNA (5'-D(P*TP*TP*GP*AP*TP*GP*GP*GP*AP*AP*TP*TP*TP*CP*CP*GP*AP*TP*TP*C)-3'), Transcription factor p65,Proto-oncogene c-Rel chimera | | Authors: | Chang, A, Wu, Y, Li, S.X, Smale, S, Chen, L. | | Deposit date: | 2023-09-19 | | Release date: | 2024-10-02 | | Method: | X-RAY DIFFRACTION (3.091 Å) | | Cite: | Crystal Structure of RelA-cRel chimera complex with DNA
To Be Published
|
|
8V9R
 
 | |
8CZA
 
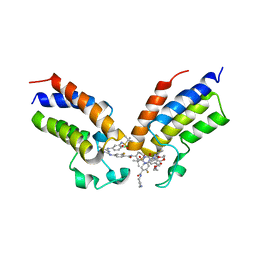 | | Crystal structure of the first bromodomain (BD1) of human BRDT bound to GXH-IV-075 | | Descriptor: | 4-[(4-{4-chloro-3-[(2-methylpropane-2-sulfonyl)amino]anilino}-5-methylpyrimidin-2-yl)amino]-2-fluoro-N-[1-(14-{3-[(2-{3-fluoro-4-[(piperidin-4-yl)carbamoyl]anilino}-5-methylpyrimidin-4-yl)amino]-5-[(2-methylpropane-2-sulfonyl)amino]phenyl}-14-oxo-4,7,10-trioxa-13-azatetradecanan-1-oyl)piperidin-4-yl]benzamide, Bromodomain testis-specific protein, SODIUM ION | | Authors: | Chan, A, Schonbrunn, E. | | Deposit date: | 2022-05-24 | | Release date: | 2022-08-03 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.96 Å) | | Cite: | Bivalent BET Bromodomain Inhibitors Confer Increased Potency and Selectivity for BRDT via Protein Conformational Plasticity.
J.Med.Chem., 65, 2022
|
|
7YC6
 
 | |
7UCS
 
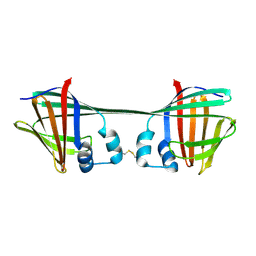 | |
7UCT
 
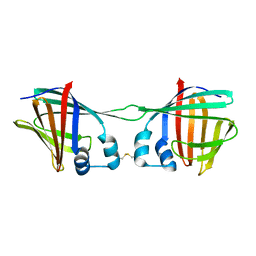 | |
7UD3
 
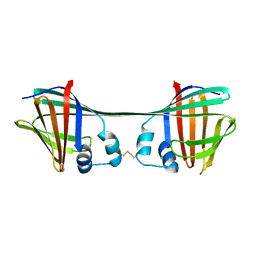 | |
7UCV
 
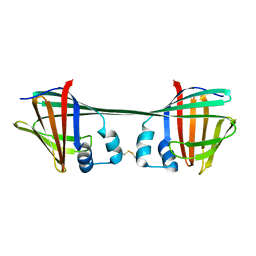 | |
7UCZ
 
 | |
7UD1
 
 | |
7UCN
 
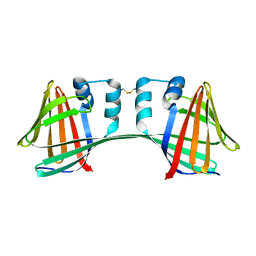 | |
1NJN
 
 | | The crystal structure of the 50S Large ribosomal subunit from Deinococcus radiodurans complexed with the antibiotic sparsomycin | | Descriptor: | 23S ribosomal RNA, SPARSOMYCIN | | Authors: | Bashan, A, Agmon, I, Zarivatch, R, Schluenzen, F, Harms, J.M, Berisio, R, Bartels, H, Hansen, H.A, Yonath, A. | | Deposit date: | 2003-01-02 | | Release date: | 2003-02-11 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Structural basis of the ribosomal machinery for Peptide bond formation,
translocation, and nascent chain progression
Mol.Cell, 11, 2003
|
|
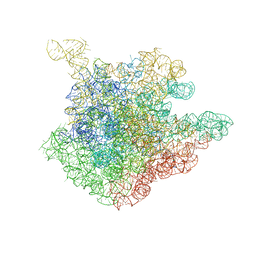
8E7V
 
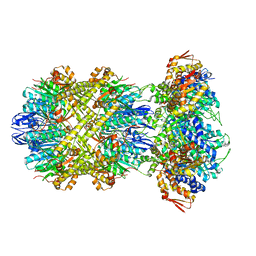 | | Cryo-EM structure of substrate-free DNClpX.ClpP from singly capped particles | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP-dependent Clp protease ATP-binding subunit ClpX, ... | | Authors: | Ghanbarpour, A, Cohen, S, Davis, J.H, Sauer, R.T. | | Deposit date: | 2022-08-24 | | Release date: | 2023-11-08 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Cryo-EM structure of substrate-free DNClpX.ClpP
Nat Commun, 2023
|
|
8E91
 
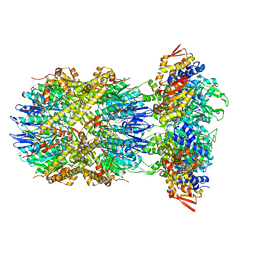 | |
8E8Q
 
 | | Cryo-EM structure of substrate-free DNClpX.ClpP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP-dependent Clp protease ATP-binding subunit ClpX, ... | | Authors: | Ghanbarpour, A, Cohen, S, Davis, J.H, Sauer, R.T. | | Deposit date: | 2022-08-25 | | Release date: | 2023-11-08 | | Method: | ELECTRON MICROSCOPY (3.12 Å) | | Cite: | Cryo-EM structure of substrate-free DNClpX.ClpP
Nat Commun, 2023
|
|
8ET3
 
 | | Cryo-EM structure of a delivery complex containing the SspB adaptor, an ssrA-tagged substrate, and the AAA+ ClpXP protease | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP-dependent Clp protease ATP-binding subunit ClpX, ATP-dependent Clp protease proteolytic subunit, ... | | Authors: | Ghanbarpour, A, Fei, X, Davis, J.H, Sauer, R.T. | | Deposit date: | 2022-10-16 | | Release date: | 2023-01-25 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | The SspB adaptor drives structural changes in the AAA+ ClpXP protease during ssrA-tagged substrate delivery.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
