8V8K
 
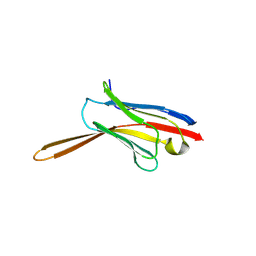 | | Crystal Structure of Nanobody NbE | | 分子名称: | Nanobody NbE | | 著者 | Koehl, A, Manglik, A, Yu, J, Kumar, A, Zhang, X, Martin, C, Raia, P, Steyaert, J, Ballet, S, Boland, A, Stoeber, M. | | 登録日 | 2023-12-05 | | 公開日 | 2024-09-11 | | 実験手法 | X-RAY DIFFRACTION (2.85 Å) | | 主引用文献 | Structural Basis of mu-Opioid Receptor-Targeting by a Nanobody Antagonist
To Be Published
|
|
5HSV
 
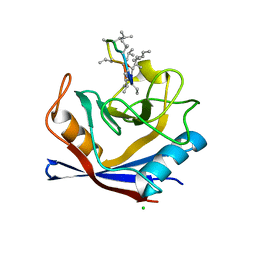 | | X-Ray structure of a CypA-Alisporivir complex at 1.5 angstrom resolution | | 分子名称: | Alisporivir, CHLORIDE ION, Peptidyl-prolyl cis-trans isomerase A | | 著者 | Dujardin, M, Bouckaert, J, Rucktooa, P, Hanoulle, X. | | 登録日 | 2016-01-26 | | 公開日 | 2017-08-16 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | X-ray structure of alisporivir in complex with cyclophilin A at 1.5 angstrom resolution.
Acta Crystallogr F Struct Biol Commun, 74, 2018
|
|
6IZK
 
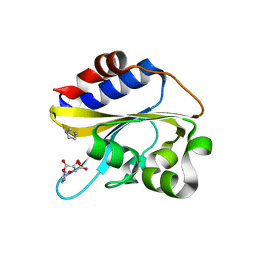 | | Structural characterization of mutated NreA protein in nitrate binding site from Staphylococcus aureus | | 分子名称: | CHLORIDE ION, IMIDAZOLE, L(+)-TARTARIC ACID, ... | | 著者 | Sangare, L, Chen, W, Wang, C, Chen, X, Wu, M, Zhang, X, Zang, J. | | 登録日 | 2018-12-19 | | 公開日 | 2020-01-22 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.29 Å) | | 主引用文献 | Structural characterization of mutated NreA protein in nitrate binding site from Staphylococcus aureus
To Be Published
|
|
177L
 
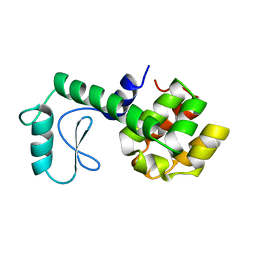 | |
226L
 
 | | GENERATING LIGAND BINDING SITES IN T4 LYSOZYME USING DEFICIENCY-CREATING SUBSTITUTIONS | | 分子名称: | BETA-MERCAPTOETHANOL, CHLORIDE ION, T4 LYSOZYME | | 著者 | Baldwin, E.P, Baase, W.A, Zhang, X.-J, Feher, V, Matthews, B.W. | | 登録日 | 1997-06-25 | | 公開日 | 1998-03-18 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Generation of ligand binding sites in T4 lysozyme by deficiency-creating substitutions.
J.Mol.Biol., 277, 1998
|
|
223L
 
 | | GENERATING LIGAND BINDING SITES IN T4 LYSOZYME USING DEFICIENCY-CREATING SUBSTITUTIONS | | 分子名称: | BENZENE, BETA-MERCAPTOETHANOL, CHLORIDE ION, ... | | 著者 | Baldwin, E.P, Baase, W.A, Zhang, X.-J, Feher, V, Matthews, B.W. | | 登録日 | 1997-06-25 | | 公開日 | 1998-03-18 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Generation of ligand binding sites in T4 lysozyme by deficiency-creating substitutions.
J.Mol.Biol., 277, 1998
|
|
7XY4
 
 | |
7XY3
 
 | |
7KDC
 
 | | The complex between RhoD and the Plexin B2 RBD | | 分子名称: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, Plexin-B2, ... | | 著者 | Kuo, Y, Wang, Y, Zhang, x. | | 登録日 | 2020-10-08 | | 公開日 | 2021-07-28 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (3.1 Å) | | 主引用文献 | A putative structural mechanism underlying the antithetic effect of homologous RND1 and RhoD GTPases in mammalian plexin regulation.
Elife, 10, 2021
|
|
6LT0
 
 | | cryo-EM structure of C9ORF72-SMCR8-WDR41 | | 分子名称: | Guanine nucleotide exchange C9orf72, Guanine nucleotide exchange protein SMCR8, WD repeat-containing protein 41 | | 著者 | Tang, D, Sheng, J, Xu, L, Zhan, X, Yan, C, Qi, S. | | 登録日 | 2020-01-21 | | 公開日 | 2020-04-15 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | Cryo-EM structure of C9ORF72-SMCR8-WDR41 reveals the role as a GAP for Rab8a and Rab11a.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
7WQV
 
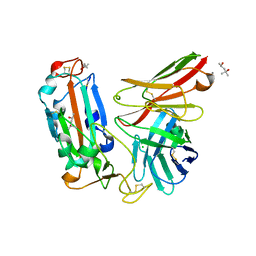 | | Crystal structure of a neutralizing monoclonal antibody (Ab08) in complex with SARS-CoV-2 receptor-binding domain (RBD) | | 分子名称: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, Ab08, ... | | 著者 | Zha, J, Meng, L, Zhang, X, Li, D. | | 登録日 | 2022-01-26 | | 公開日 | 2023-01-25 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | A Spike-destructing human antibody effectively neutralizes Omicron-included SARS-CoV-2 variants with therapeutic efficacy.
Plos Pathog., 19, 2023
|
|
8WRB
 
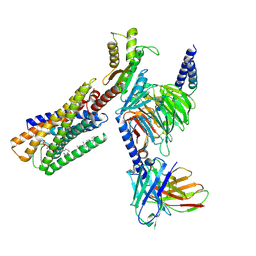 | | Lysophosphatidylserine receptor GPR34-Gi complex | | 分子名称: | Antibody fragment scFv16, CHOLESTEROL, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | 著者 | Gong, W, Liu, G, Li, X, Wang, Y, Zhang, X. | | 登録日 | 2023-10-13 | | 公開日 | 2023-11-08 | | 最終更新日 | 2023-12-20 | | 実験手法 | ELECTRON MICROSCOPY (2.91 Å) | | 主引用文献 | Structural basis for ligand recognition and signaling of the lysophosphatidylserine receptors GPR34 and GPR174.
Plos Biol., 21, 2023
|
|
167L
 
 | |
180L
 
 | |
8WOJ
 
 | |
178L
 
 | | Protein flexibility and adaptability seen in 25 crystal forms of T4 LYSOZYME | | 分子名称: | CHLORIDE ION, T4 LYSOZYME | | 著者 | Matsumura, M, Weaver, L, Zhang, X.-J, Matthews, B.W. | | 登録日 | 1995-03-24 | | 公開日 | 1995-07-10 | | 最終更新日 | 2021-11-03 | | 実験手法 | X-RAY DIFFRACTION (2.71 Å) | | 主引用文献 | Protein flexibility and adaptability seen in 25 crystal forms of T4 lysozyme.
J.Mol.Biol., 250, 1995
|
|
8WOQ
 
 | | Cryo-EM structure of human SIDT1 protein with C1 symmetry at neutral pH | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | 著者 | Liu, W, Tang, M, Wang, J, Zhang, X, Wu, S, Ru, H. | | 登録日 | 2023-10-07 | | 公開日 | 2024-03-20 | | 実験手法 | ELECTRON MICROSCOPY (2.85 Å) | | 主引用文献 | Structural insights into cholesterol transport and hydrolase activity of a putative human RNA transport protein SIDT1.
Cell Discov, 10, 2024
|
|
8WOR
 
 | | Cryo-EM structure of human SIDT1 protein with C2 symmetry at neutral pH | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | 著者 | Liu, W, Tang, M, Wang, J, Zhang, X, Wu, S, Ru, H. | | 登録日 | 2023-10-07 | | 公開日 | 2024-03-20 | | 実験手法 | ELECTRON MICROSCOPY (2.66 Å) | | 主引用文献 | Structural insights into cholesterol transport and hydrolase activity of a putative human RNA transport protein SIDT1.
Cell Discov, 10, 2024
|
|
172L
 
 | |
4HBK
 
 | | Structure of the Aldose Reductase from Schistosoma japonicum | | 分子名称: | Aldo-keto reductase family 1, member B4 (Aldose reductase) | | 著者 | Liu, J, Cheng, J, Zhang, X, Yang, Z, Hu, W, Xu, Y. | | 登録日 | 2012-09-28 | | 公開日 | 2013-06-26 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Aldose reductase from Schistosoma japonicum: crystallization and structure-based inhibitor screening for discovering antischistosomal lead compounds.
Parasit Vectors, 6, 2013
|
|
2B37
 
 | | Crystal structure of Mycobacterium tuberculosis enoyl reductase (InhA) inhibited by 5-octyl-2-phenoxyphenol | | 分子名称: | 5-OCTYL-2-PHENOXYPHENOL, Enoyl-[acyl-carrier-protein] reductase [NADH], NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | 著者 | Sullivan, T.J, Truglio, J.J, Novichenok, P, Stratton, C, Zhang, X, Kaur, T, Johnson, F, Boyne, M.S, Amin, A. | | 登録日 | 2005-09-19 | | 公開日 | 2006-03-07 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | High Affinity InhA Inhibitors with Activity against Drug-Resistant Strains
of Mycobacterium tuberculosis
ACS Chem.Biol., 1, 2006
|
|
7FIK
 
 | | The cryo-EM structure of the CR subunit from X. laevis NPC | | 分子名称: | MGC154553 protein, MGC83295 protein, MGC83926 protein, ... | | 著者 | Shi, Y, Huang, G, Zhan, X. | | 登録日 | 2021-07-31 | | 公開日 | 2022-11-09 | | 最終更新日 | 2024-06-12 | | 実験手法 | ELECTRON MICROSCOPY (3.7 Å) | | 主引用文献 | Structure of the cytoplasmic ring of the Xenopus laevis nuclear pore complex.
Science, 376, 2022
|
|
2GIC
 
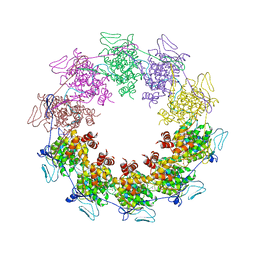 | | Crystal Structure of a vesicular stomatitis virus nucleocapsid-RNA complex | | 分子名称: | 45-MER, Nucleocapsid protein, URANYL (VI) ION | | 著者 | Green, T.J, Zhang, X, Wertz, G.W, Luo, M. | | 登録日 | 2006-03-28 | | 公開日 | 2006-08-22 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (2.92 Å) | | 主引用文献 | Structure of the vesicular stomatitis virus nucleoprotein-RNA complex unveils how the RNA is sequestered
Science, 313, 2006
|
|
2KA9
 
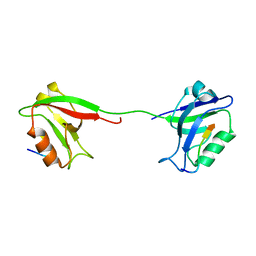 | | Solution structure of PSD-95 PDZ12 complexed with cypin peptide | | 分子名称: | Disks large homolog 4, cypin peptide | | 著者 | Wang, W.N, Weng, J.W, Zhang, X, Liu, M.L, Zhang, M.J. | | 登録日 | 2008-11-03 | | 公開日 | 2009-06-23 | | 最終更新日 | 2024-05-29 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Creating conformational entropy by increasing interdomain mobility in ligand binding regulation: a revisit to N-terminal tandem PDZ domains of PSD-95
J.Am.Chem.Soc., 131, 2009
|
|
2IBJ
 
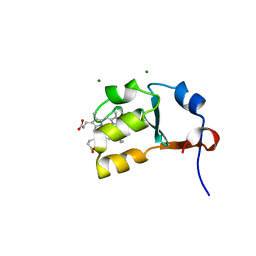 | | Structure of House Fly Cytochrome B5 | | 分子名称: | Cytochrome b5, MAGNESIUM ION, PROTOPORPHYRIN IX CONTAINING FE | | 著者 | Terzyan, S, Zhang, X.C, Benson, D.R, Wang, L. | | 登録日 | 2006-09-11 | | 公開日 | 2007-02-27 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (1.55 Å) | | 主引用文献 | Comparison of cytochromes b(5) from insects and vertebrates.
Proteins, 67, 2007
|
|
