1F4K
 
 | | CRYSTAL STRUCTURE OF THE REPLICATION TERMINATOR PROTEIN/B-SITE DNA COMPLEX | | Descriptor: | 5'-D(*CP*TP*AP*TP*GP*AP*AP*CP*AP*TP*AP*AP*TP*GP*TP*TP*CP*AP*TP*AP*G)-3', 5'-D(*CP*TP*AP*TP*GP*AP*AP*CP*AP*TP*TP*AP*TP*GP*TP*TP*CP*AP*TP*AP*G)-3', REPLICATION TERMINATION PROTEIN | | Authors: | Wilce, J.A, Vivian, J.P, Hastings, A.F, Otting, G, Folmer, R.H.A, Duggin, I.G, Wake, R.G, Wilce, M.C.J. | | Deposit date: | 2000-06-08 | | Release date: | 2001-06-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of the RTP-DNA complex and the mechanism of polar replication fork arrest
Nat.Struct.Biol., 8, 2001
|
|
1F5O
 
 | |
2PZE
 
 | | Minimal human CFTR first nucleotide binding domain as a head-to-tail dimer | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Cystic fibrosis transmembrane conductance regulator, MAGNESIUM ION | | Authors: | Atwell, S, Conners, K, Emtage, S, Gheyi, T, Glenn, N.R, Hendle, J, Lewis, H.A, Lu, F, Rodgers, L.A, Romero, R, Sauder, J.M, Smith, D, Tien, H, Wasserman, S.R, Zhao, X. | | Deposit date: | 2007-05-17 | | Release date: | 2007-10-09 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structures of a minimal human CFTR first nucleotide-binding domain as a monomer, head-to-tail homodimer, and pathogenic mutant.
Protein Eng.Des.Sel., 23, 2010
|
|
1F5P
 
 | |
2PZF
 
 | | Minimal human CFTR first nucleotide binding domain as a head-to-tail dimer with delta F508 | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Cystic fibrosis transmembrane conductance regulator, MAGNESIUM ION | | Authors: | Atwell, S, Conners, K, Emtage, S, Gheyi, T, Glenn, N.R, Hendle, J, Lewis, H.A, Lu, F, Rodgers, L.A, Romero, R, Sauder, J.M, Smith, D, Tien, H, Wasserman, S.R, Zhao, X. | | Deposit date: | 2007-05-18 | | Release date: | 2007-10-09 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structures of a minimal human CFTR first nucleotide-binding domain as a monomer, head-to-tail homodimer, and pathogenic mutant.
Protein Eng.Des.Sel., 23, 2010
|
|
2N4U
 
 | |
2N4R
 
 | |
2PNJ
 
 | | Crystal structure of human ferrochelatase mutant with Phe 337 replaced by Ala | | Descriptor: | CHOLIC ACID, FE2/S2 (INORGANIC) CLUSTER, Ferrochelatase, ... | | Authors: | Dailey, H.A, Wu, C.-K, Horanyi, P, Medlock, A.E, Najahi-Missaoui, W, Burden, A.E, Dailey, T.A, Rose, J.P. | | Deposit date: | 2007-04-24 | | Release date: | 2007-10-02 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Altered orientation of active site residues in variants of human ferrochelatase. Evidence for a hydrogen bond network involved in catalysis
Biochemistry, 46, 2007
|
|
2QD3
 
 | | Wild type human ferrochelatase crystallized with ammonium sulfate | | Descriptor: | CHOLIC ACID, FE2/S2 (INORGANIC) CLUSTER, Ferrochelatase, ... | | Authors: | Medlock, A.E, Dailey, T.A, Ross, T.A, Dailey, H.A, Lanzilotta, W.N. | | Deposit date: | 2007-06-20 | | Release date: | 2007-10-30 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | A pi-Helix Switch Selective for Porphyrin Deprotonation and Product Release in Human Ferrochelatase.
J.Mol.Biol., 373, 2007
|
|
8QOD
 
 | | CRYSTAL STRUCTURE OF FVIIA IN COMPLEX WITH A BENZAMIDINE-BASED INHIBITOR | | Descriptor: | 3-[2-bromanyl-4-[(1~{S})-1-[[3-[(4-carbamimidoylphenyl)amino]-3-oxidanylidene-propanoyl]amino]ethyl]phenoxy]benzoic acid, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Schreuder, H.A, Liesum, A. | | Deposit date: | 2023-09-28 | | Release date: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.573 Å) | | Cite: | Crystallography and molecular simulations capture S1 pocket
collapse in allosteric regulation of factor VIIa and other serine
proteases.
To be published
|
|
1I4W
 
 | | THE CRYSTAL STRUCTURE OF THE TRANSCRIPTION FACTOR SC-MTTFB OFFERS INTRIGUING INSIGHTS INTO MITOCHONDRIAL TRANSCRIPTION | | Descriptor: | MITOCHONDRIAL REPLICATION PROTEIN MTF1, XENON | | Authors: | Schubot, F.D, Chen, C.-J, Rose, J.P, Dailey, T.A, Dailey, H.A, Wang, B.-C. | | Deposit date: | 2001-02-23 | | Release date: | 2001-10-03 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of the transcription factor sc-mtTFB offers insights into mitochondrial transcription.
Protein Sci., 10, 2001
|
|
2HRC
 
 | | 1.7 angstrom structure of human ferrochelatase variant R115L | | Descriptor: | CHLORIDE ION, CHOLIC ACID, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Medlock, A, Swartz, L, Dailey, T.A, Dailey, H.A, Lanzilotta, W.N. | | Deposit date: | 2006-07-20 | | Release date: | 2007-03-13 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Substrate interactions with human ferrochelatase
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
2R69
 
 | | Crystal structure of Fab 1A1D-2 complexed with E-DIII of Dengue virus at 3.8 angstrom resolution | | Descriptor: | Heavy chain of 1A1D-2, Light chain of 1A1D-2, Major envelope protein E | | Authors: | Lok, S.M, Kostyuchenko, V.K, Nybakken, G.E, Holdaway, H.A, Battisti, A.J, Sukupolvi-petty, S, Sedlak, D, Fremont, D.H, Chipman, P.R, Roehrig, J.T, Diamond, M.S, Kuhn, R.J, Rossmann, M.G. | | Deposit date: | 2007-09-05 | | Release date: | 2007-12-25 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Binding of a neutralizing antibody to dengue virus alters the arrangement of surface glycoproteins.
Nat.Struct.Mol.Biol., 15, 2008
|
|
2HRE
 
 | | Structure of human ferrochelatase variant E343K with protoporphyrin IX bound | | Descriptor: | CHOLIC ACID, FE2/S2 (INORGANIC) CLUSTER, Ferrochelatase, ... | | Authors: | Medlock, A, Swartz, L, Dailey, T.A, Dailey, H.A, Lanzilotta, W.N. | | Deposit date: | 2006-07-20 | | Release date: | 2007-03-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Substrate interactions with human ferrochelatase
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
1A5Q
 
 | | P93A VARIANT OF BOVINE PANCREATIC RIBONUCLEASE A | | Descriptor: | RIBONUCLEASE A, SULFATE ION | | Authors: | Pearson, M.A, Karplus, P.A, Dodge, R.W, Laity, J.H, Scheraga, H.A. | | Deposit date: | 1998-02-17 | | Release date: | 1998-05-27 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structures of two mutants that have implications for the folding of bovine pancreatic ribonuclease A.
Protein Sci., 7, 1998
|
|
1A60
 
 | | NMR STRUCTURE OF A CLASSICAL PSEUDOKNOT: INTERPLAY OF SINGLE-AND DOUBLE-STRANDED RNA, 24 STRUCTURES | | Descriptor: | TYMV PSEUDOKNOT | | Authors: | Kolk, M.H, Van Der Graaf, M, Wijmenga, S.S, Pleij, C.W.A, Heus, H.A, Hilbers, C.W. | | Deposit date: | 1998-03-04 | | Release date: | 1998-05-27 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | NMR structure of a classical pseudoknot: interplay of single- and double-stranded RNA.
Science, 280, 1998
|
|
2R6P
 
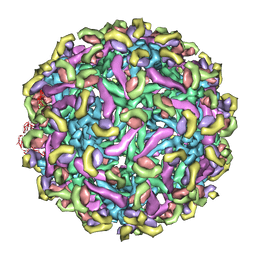 | | Fit of E protein and Fab 1A1D-2 into 24 angstrom resolution cryoEM map of Fab complexed with dengue 2 virus. | | Descriptor: | Heavy chain of 1A1D-2, Light chain of 1A1D-2, Major envelope protein E | | Authors: | Lok, S.M, Kostyuchenko, V.K, Holdaway, H.A, Chipman, P.R, Kuhn, R.J, Rossmann, M.G. | | Deposit date: | 2007-09-06 | | Release date: | 2007-12-25 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (24 Å) | | Cite: | Binding of a neutralizing antibody to dengue virus alters the arrangement of surface glycoproteins.
Nat.Struct.Mol.Biol., 15, 2008
|
|
2IL6
 
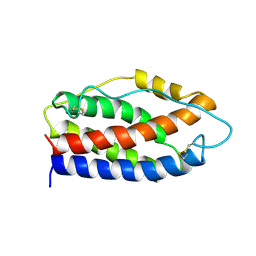 | | HUMAN INTERLEUKIN-6, NMR, 32 STRUCTURES | | Descriptor: | INTERLEUKIN-6 | | Authors: | Xu, G.Y, Yu, H.A, Hong, J, Stahl, M, Mcdonagh, T, Kay, L.E, Cumming, D.A. | | Deposit date: | 1997-01-31 | | Release date: | 1998-02-04 | | Last modified: | 2022-03-09 | | Method: | SOLUTION NMR | | Cite: | Solution structure of recombinant human interleukin-6.
J.Mol.Biol., 268, 1997
|
|
2J4I
 
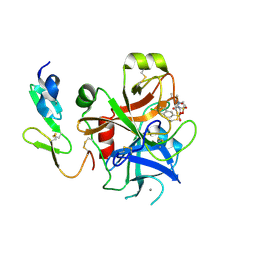 | | CRYSTAL STRUCTURE OF A HUMAN FACTOR XA INHIBITOR COMPLEX | | Descriptor: | 1-PYRROLIDINEACETAMIDE, 3-[[(6-CHLORO-2-NAPHTHALENYL)SULFONYL]AMINO]-ALPHA-METHYL-N-(1-METHYLETHYL)-N-[2-[(METHYLSULFONYL)AMINO]ETHYL]-2-OXO-, (ALPHAS,3S)-, ... | | Authors: | Young, R.J, Campbell, M, Borthwick, A.D, Brown, D, Chan, C, Convery, M.A, Crowe, M.C, Dayal, S, Diallo, H, Kelly, H.A, Paul King, N, Kleanthous, S, Kurtis, C.L, Mason, A.M, Mordaunt, J.E, Patel, C, Pateman, A.J, Senger, S, Shah, G.P, Smith, P.W, Watson, N.S, Weston, H.E, Zhou, P. | | Deposit date: | 2006-08-31 | | Release date: | 2006-09-27 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure- and Property-Based Design of Factor Xa Inhibitors: Pyrrolidin-2-Ones with Acyclic Alanyl Amides as P4 Motifs.
Bioorg.Med.Chem.Lett., 16, 2006
|
|
1IHF
 
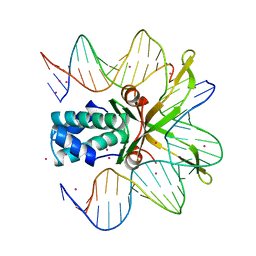 | | INTEGRATION HOST FACTOR/DNA COMPLEX | | Descriptor: | CADMIUM ION, DNA (35-MER), DNA (5'-D(*GP*CP*TP*TP*AP*TP*CP*AP*AP*TP*TP*TP*GP*TP*TP*GP*C P*AP*CP*C)-3'), ... | | Authors: | Rice, P.A, Yang, S.-W, Mizuuchi, K, Nash, H.A. | | Deposit date: | 1996-08-21 | | Release date: | 1997-02-12 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of an IHF-DNA complex: a protein-induced DNA U-turn.
Cell(Cambridge,Mass.), 87, 1996
|
|
1B0E
 
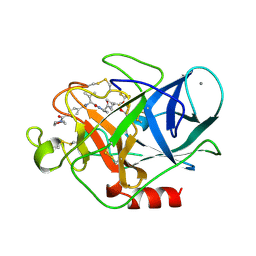 | | CRYSTAL STRUCTURE OF PORCINE PANCREATIC ELASTASE WITH MDL 101,146 | | Descriptor: | 1-{3-METHYL-2-[4-(MORPHOLINE-4-CARBONYL)-BENZOYLAMINO]-BUTYRYL}-PYRROLIDINE-2-CARBOXYLIC ACID (3,3,4,4,4-PENTAFLUORO-1-ISOPROPYL-2-OXO-BUTYL)-AMIDE, CALCIUM ION, PROTEIN (ELASTASE) | | Authors: | Schreuder, H.A, Metz, W.A, Peet, N.P, Pelton, J.T, Tardif, C. | | Deposit date: | 1998-11-09 | | Release date: | 1998-11-18 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Inhibition of human neutrophil elastase. 4. Design, synthesis, X-ray crystallographic analysis, and structure-activity relationships for a series of P2-modified, orally active peptidyl pentafluoroethyl ketones.
J.Med.Chem., 41, 1998
|
|
1ATO
 
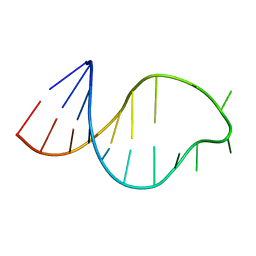 | | THE STRUCTURE OF THE ISOLATED, CENTRAL HAIRPIN OF THE HDV ANTIGENOMIC RIBOZYME, NMR, 10 STRUCTURES | | Descriptor: | RNA (5'-R(*GP*GP*CP*AP*CP*CP*UP*CP*CP*UP*CP*GP*CP*GP*GP*UP*GP*CP*C)-3') | | Authors: | Kolk, M.H, Heus, H.A, Hilbers, C.W. | | Deposit date: | 1997-08-14 | | Release date: | 1997-11-12 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The structure of the isolated, central hairpin of the HDV antigenomic ribozyme: novel structural features and similarity of the loop in the ribozyme and free in solution.
EMBO J., 16, 1997
|
|
1J6X
 
 | | CRYSTAL STRUCTURE OF HELICOBACTER PYLORI LUXS | | Descriptor: | AUTOINDUCER-2 PRODUCTION PROTEIN LUXS, METHIONINE, ZINC ION | | Authors: | Lewis, H.A, Furlong, E.B, Bergseid, M.G, Sanderson, W.E, Buchanan, S.G. | | Deposit date: | 2001-05-14 | | Release date: | 2001-06-08 | | Last modified: | 2017-10-04 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | A structural genomics approach to the study of quorum sensing: crystal structures of three LuxS orthologs.
Structure, 9, 2001
|
|
2J94
 
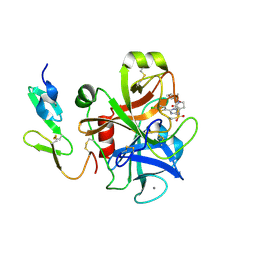 | | CRYSTAL STRUCTURE OF A HUMAN FACTOR XA INHIBITOR COMPLEX | | Descriptor: | 5-(5-CHLORO-2-THIENYL)-N-{(3S)-1-[(1S)-1-METHYL-2-MORPHOLIN-4-YL-2-OXOETHYL]-2-OXOPYRROLIDIN-3-YL}-1H-1,2,4-TRIAZOLE-3-SULFONAMIDE, CALCIUM ION, COAGULATION FACTOR X | | Authors: | Chan, C, Borthwick, A.D, Brown, D, Campbell, M, Chaudry, L, Chung, C.W, Convery, M.A, Hamblin, J.N, Johnstone, L, Kelly, H.A, Kleanthous, S, Burns-Kurtis, C.L, Patikis, A, Patel, C, Pateman, A.J, Senger, S, Shah, G.P, Toomey, J.R, Watson, N.S, Weston, H.E, Whitworth, C, Young, R.J, Zhou, P. | | Deposit date: | 2006-11-02 | | Release date: | 2007-03-20 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Factor Xa Inhibitors: S1 Binding Interactions of a Series of N-{(3S)-1-[(1S)-1-Methyl-2-Morpholin-4-Yl-2-Oxoethyl]-2-Oxopyrrolidin-3-Yl}Sulfonamides.
J.Med.Chem., 50, 2007
|
|
1IRA
 
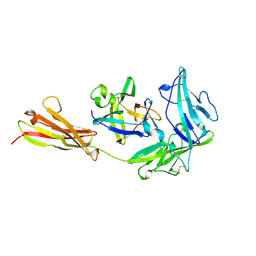 | | COMPLEX OF THE INTERLEUKIN-1 RECEPTOR WITH THE INTERLEUKIN-1 RECEPTOR ANTAGONIST (IL1RA) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, INTERLEUKIN-1 RECEPTOR, INTERLEUKIN-1 RECEPTOR ANTAGONIST | | Authors: | Schreuder, H.A, Tardif, C, Tramp-Kalmeyer, S, Soffientini, A, Sarubbi, E, Akeson, A, Bowlin, T, Yanofsky, S, Barrett, R.W. | | Deposit date: | 1998-04-09 | | Release date: | 1998-06-17 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | A new cytokine-receptor binding mode revealed by the crystal structure of the IL-1 receptor with an antagonist.
Nature, 386, 1997
|
|
