6QTM
 
 | | Crystal structure of the Sir4 H-BRCT domain in complex with Ty5 pS1095 peptide | | 分子名称: | Regulatory protein SIR4, Ribonuclease H, SULFATE ION | | 著者 | Gut, H, Deshpande, I, Keusch, J.J, Challa, K, Iesmantavicius, V, Gasser, S.M. | | 登録日 | 2019-02-25 | | 公開日 | 2019-09-18 | | 最終更新日 | 2021-08-11 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | The Sir4 H-BRCT domain interacts with phospho-proteins to sequester and repress yeast heterochromatin.
Embo J., 38, 2019
|
|
3HBX
 
 | | Crystal structure of GAD1 from Arabidopsis thaliana | | 分子名称: | Glutamate decarboxylase 1 | | 著者 | Gut, H, Dominici, P, Pilati, S, Gruetter, M.G, Capitani, G. | | 登録日 | 2009-05-05 | | 公開日 | 2009-07-28 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.672 Å) | | 主引用文献 | A common structural basis for pH- and calmodulin-mediated regulation in plant glutamate decarboxylase.
J.Mol.Biol., 392, 2009
|
|
2JKB
 
 | | Crystal structure of Streptococcus pneumoniae NanB in complex with 2, 7-anhydro-Neu5Ac | | 分子名称: | 1,2-ETHANEDIOL, 2-ACETYLAMINO-7-(1,2-DIHYDROXY-ETHYL)-3-HYDROXY-6,8-DIOXA-BICYCLO[3.2.1]OCTANE-5-CARBOXYLIC ACID, SIALIDASE B | | 著者 | Gut, H, King, S.J, Walsh, M.A. | | 登録日 | 2008-08-26 | | 公開日 | 2008-09-16 | | 最終更新日 | 2024-05-01 | | 実験手法 | X-RAY DIFFRACTION (1.54 Å) | | 主引用文献 | Structural and Functional Studies of Streptococcus Pneumoniae Neuraminidase B: An Intramolecular Trans-Sialidase.
FEBS Lett., 582, 2008
|
|
2YA6
 
 | | Crystal structure of Streptococcus pneumoniae NanA (TIGR4) in complex with DANA | | 分子名称: | 2-DEOXY-2,3-DEHYDRO-N-ACETYL-NEURAMINIC ACID, CHLORIDE ION, FORMIC ACID, ... | | 著者 | Gut, H, Xu, G, Taylor, G.L, Walsh, M.A. | | 登録日 | 2011-02-18 | | 公開日 | 2011-04-27 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structural Basis for Streptococcus Pneumoniae Nana Inhibition by Influenza Antivirals Zanamivir and Oseltamivir Carboxylate.
J.Mol.Biol., 409, 2011
|
|
2YA8
 
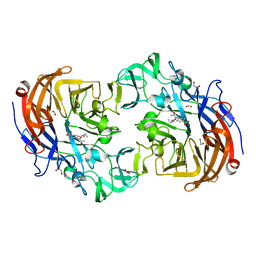 | | Crystal structure of Streptococcus pneumoniae NanA (TIGR4) in complex with Oseltamivir carboxylate | | 分子名称: | (3R,4R,5S)-4-(acetylamino)-5-amino-3-(pentan-3-yloxy)cyclohex-1-ene-1-carboxylic acid, 1,2-ETHANEDIOL, CHLORIDE ION, ... | | 著者 | Gut, H, Xu, G, Taylor, G.L, Walsh, M.A. | | 登録日 | 2011-02-18 | | 公開日 | 2011-04-27 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | Structural Basis for Streptococcus Pneumoniae Nana Inhibition by Influenza Antivirals Zanamivir and Oseltamivir Carboxylate.
J.Mol.Biol., 409, 2011
|
|
2YA5
 
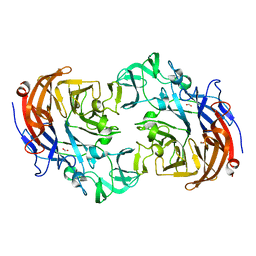 | | Crystal structure of Streptococcus pneumoniae NanA (TIGR4) in complex with sialic acid | | 分子名称: | CHLORIDE ION, FORMIC ACID, N-acetyl-alpha-neuraminic acid, ... | | 著者 | Gut, H, Xu, G, Taylor, G.L, Walsh, M.A. | | 登録日 | 2011-02-18 | | 公開日 | 2011-04-27 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structural Basis for Streptococcus Pneumoniae Nana Inhibition by Influenza Antivirals Zanamivir and Oseltamivir Carboxylate.
J.Mol.Biol., 409, 2011
|
|
2YA7
 
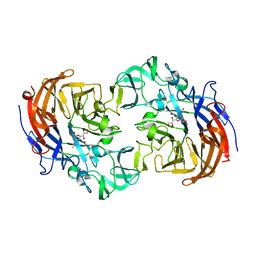 | | Crystal structure of Streptococcus pneumoniae NanA (TIGR4) in complex with Zanamivir | | 分子名称: | CHLORIDE ION, NEURAMINIDASE A, ZANAMIVIR | | 著者 | Gut, H, Xu, G, Taylor, G.L, Walsh, M.A. | | 登録日 | 2011-02-18 | | 公開日 | 2011-04-27 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (1.89 Å) | | 主引用文献 | Structural Basis for Streptococcus Pneumoniae Nana Inhibition by Influenza Antivirals Zanamivir and Oseltamivir Carboxylate.
J.Mol.Biol., 409, 2011
|
|
2YA4
 
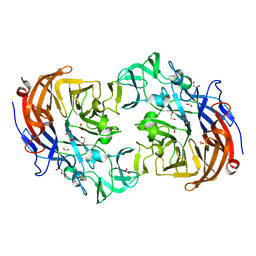 | | Crystal structure of Streptococcus pneumoniae NanA (TIGR4) | | 分子名称: | 1,2-ETHANEDIOL, CHLORIDE ION, FORMIC ACID, ... | | 著者 | Gut, H, Xu, G, Taylor, G.L, Walsh, M.A. | | 登録日 | 2011-02-18 | | 公開日 | 2011-04-27 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Structural Basis for Streptococcus Pneumoniae Nana Inhibition by Influenza Antivirals Zanamivir and Oseltamivir Carboxylate.
J.Mol.Biol., 409, 2011
|
|
1M7Y
 
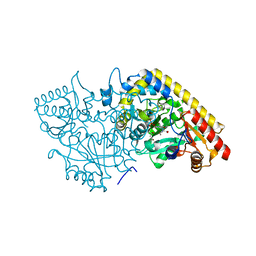 | | Crystal structure of apple ACC synthase in complex with L-aminoethoxyvinylglycine | | 分子名称: | (2E,3E)-4-(2-aminoethoxy)-2-[({3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methyl)imino]but-3-enoic acid, (4R)-2-METHYLPENTANE-2,4-DIOL, 1-aminocyclopropane-1-carboxylate synthase | | 著者 | Capitani, G, McCarthy, D, Gut, H, Gruetter, M.G, Kirsch, J.F. | | 登録日 | 2002-07-23 | | 公開日 | 2002-12-23 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Apple 1-Aminocyclopropane-1-carboxylate Synthase in Complex with the Inhibitor
L-Aminoethoxyvinylglycine
J.Biol.Chem., 277, 2002
|
|
1M4N
 
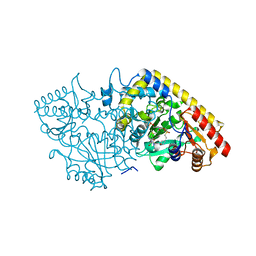 | | CRYSTAL STRUCTURE OF APPLE ACC SYNTHASE IN COMPLEX WITH [2-(AMINO-OXY)ETHYL](5'-DEOXYADENOSIN-5'-YL)(METHYL)SULFONIUM | | 分子名称: | (2-AMINOOXY-ETHYL)-[5-(6-AMINO-PURIN-9-YL)-3,4-DIHYDROXY-TETRAHYDRO-FURAN-2-YLMETHYL]-METHYL-SULFONIUM, 1-aminocyclopropane-1-carboxylate synthase, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ... | | 著者 | Capitani, G, Eliot, A.C, Gut, H, Khomutov, R.M, Kirsch, J.F, Grutter, M.G. | | 登録日 | 2002-07-03 | | 公開日 | 2003-04-22 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.01 Å) | | 主引用文献 | Structure of 1-aminocyclopropane-1-carboxylate synthase in complex with an amino-oxy analogue of the substrate: implications for substrate binding.
BIOCHEM.BIOPHYS.ACTA PROTEINS & PROTEOMICS, 1647, 2003
|
|
6QSZ
 
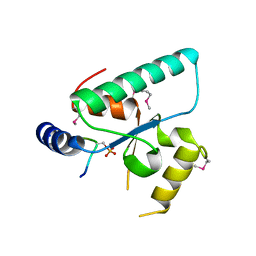 | | Crystal structure of the Sir4 H-BRCT domain in complex with Esc1 pS1450 peptide | | 分子名称: | CHLORIDE ION, Regulatory protein SIR4, Silent chromatin protein ESC1 | | 著者 | Deshpande, I, Keusch, J.J, Challa, K, Iesmantavicius, V, Gasser, S.M, Gut, H. | | 登録日 | 2019-02-22 | | 公開日 | 2019-09-18 | | 最終更新日 | 2019-10-23 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | The Sir4 H-BRCT domain interacts with phospho-proteins to sequester and repress yeast heterochromatin.
Embo J., 38, 2019
|
|
4UMG
 
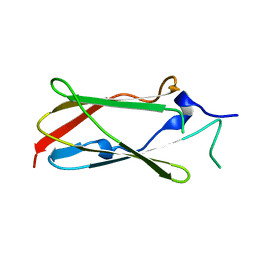 | | Crystal structure of the Lin-41 filamin domain | | 分子名称: | PROTEIN LIN-41 | | 著者 | Tocchini, C, Keusch, J.J, Miller, S.B, Finger, S, Gut, H, Stadler, M, Ciosk, R. | | 登録日 | 2014-05-16 | | 公開日 | 2014-10-01 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (1.68 Å) | | 主引用文献 | The Trim-Nhl Protein Lin-41 Controls the Onset of Developmental Plasticity in Caenorhabditis Elegans.
Plos Genet., 10, 2014
|
|
1PMM
 
 | | Crystal structure of Escherichia coli GadB (low pH) | | 分子名称: | ACETIC ACID, Glutamate decarboxylase beta, PYRIDOXAL-5'-PHOSPHATE | | 著者 | Capitani, G, De Biase, D, Aurizi, C, Gut, H, Bossa, F, Grutter, M.G. | | 登録日 | 2003-06-11 | | 公開日 | 2004-02-17 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Crystal structure and functional analysis of escherichia coli glutamate
decarboxylase
Embo J., 22, 2003
|
|
1PMO
 
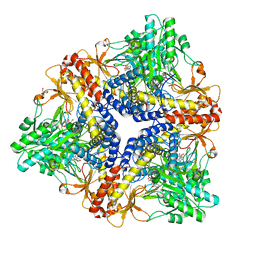 | | Crystal structure of Escherichia coli GadB (neutral pH) | | 分子名称: | (5-HYDROXY-4,6-DIMETHYLPYRIDIN-3-YL)METHYL DIHYDROGEN PHOSPHATE, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Glutamate decarboxylase beta | | 著者 | Capitani, G, De Biase, D, Aurizi, C, Gut, H, Bossa, F, Grutter, M.G. | | 登録日 | 2003-06-11 | | 公開日 | 2004-02-17 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Crystal structure and functional analysis of escherichia coli glutamate
decarboxylase
Embo J., 22, 2003
|
|
2DGL
 
 | | Crystal structure of Escherichia coli GadB in complex with bromide | | 分子名称: | ACETIC ACID, BROMIDE ION, Glutamate decarboxylase beta, ... | | 著者 | Gruetter, M.G, Capitani, G, Gut, H. | | 登録日 | 2006-03-14 | | 公開日 | 2006-06-20 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (3.15 Å) | | 主引用文献 | Escherichia coli acid resistance: pH-sensing, activation by chloride and autoinhibition in GadB
Embo J., 25, 2006
|
|
2DGM
 
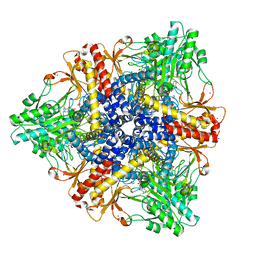 | | Crystal structure of Escherichia coli GadB in complex with iodide | | 分子名称: | ACETIC ACID, DI(HYDROXYETHYL)ETHER, FORMIC ACID, ... | | 著者 | Gruetter, M.G, Capitani, G, Gut, H. | | 登録日 | 2006-03-14 | | 公開日 | 2006-06-20 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Escherichia coli acid resistance: pH-sensing, activation by chloride and autoinhibition in GadB
Embo J., 25, 2006
|
|
2DGK
 
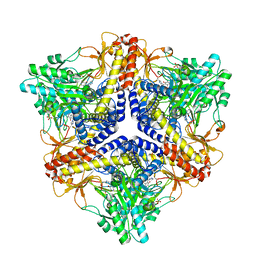 | | Crystal structure of an N-terminal deletion mutant of Escherichia coli GadB in an autoinhibited state (aldamine) | | 分子名称: | 1,2-ETHANEDIOL, Glutamate decarboxylase beta, PYRIDOXAL-5'-PHOSPHATE, ... | | 著者 | Gruetter, M.G, Capitani, G, Gut, H. | | 登録日 | 2006-03-14 | | 公開日 | 2006-06-20 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Escherichia coli acid resistance: pH-sensing, activation by chloride and autoinhibition in GadB
Embo J., 25, 2006
|
|
2YBA
 
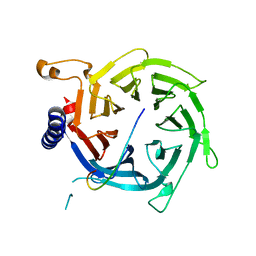 | | Crystal structure of Nurf55 in complex with histone H3 | | 分子名称: | HISTONE H3, PROBABLE HISTONE-BINDING PROTEIN CAF1 | | 著者 | Schmitges, F.W, Prusty, A.B, Faty, M, Stutzer, A, Lingaraju, G.M, Aiwazian, J, Sack, R, Hess, D, Li, L, Zhou, S, Bunker, R.D, Wirth, U, Bouwmeester, T, Bauer, A, Ly-Hartig, N, Zhao, K, Chan, H, Gu, J, Gut, H, Fischle, W, Muller, J, Thoma, N.H. | | 登録日 | 2011-03-02 | | 公開日 | 2011-05-11 | | 最終更新日 | 2024-05-01 | | 実験手法 | X-RAY DIFFRACTION (2.55 Å) | | 主引用文献 | Histone Methylation by Prc2 is Inhibited by Active Chromatin Marks
Mol.Cell, 42, 2011
|
|
2YB8
 
 | | Crystal structure of Nurf55 in complex with Su(z)12 | | 分子名称: | POLYCOMB PROTEIN SU(Z)12, PROBABLE HISTONE-BINDING PROTEIN CAF1, SULFATE ION | | 著者 | Schmitges, F.W, Prusty, A.B, Faty, M, Stutzer, A, Lingaraju, G.M, Aiwazian, J, Sack, R, Hess, D, Li, L, Zhou, S, Bunker, R.D, Wirth, U, Bouwmeester, T, Bauer, A, Ly-Hartig, N, Zhao, K, Chan, H, Gu, J, Gut, H, Fischle, W, Muller, J, Thoma, N.H. | | 登録日 | 2011-03-02 | | 公開日 | 2011-05-18 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Histone Methylation by Prc2 is Inhibited by Active Chromatin Marks.
Mol.Cell, 42, 2011
|
|
5OMD
 
 | | Crystal structure of S. cerevisiae Ddc2 N-terminal coiled-coil domain | | 分子名称: | DNA damage checkpoint protein LCD1 | | 著者 | Deshpande, I, Seeber, A, Shimada, K, Keusch, J.J, Gut, H, Gasser, S.M. | | 登録日 | 2017-07-28 | | 公開日 | 2017-10-25 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Structural Basis of Mec1-Ddc2-RPA Assembly and Activation on Single-Stranded DNA at Sites of Damage.
Mol. Cell, 68, 2017
|
|
5OMC
 
 | | Crystal structure of K. lactis Ddc2 N-terminus in complex with S. cerevisiae Rfa1 (K45E mutant) N-OB domain | | 分子名称: | CHLORIDE ION, DNA damage checkpoint protein LCD1, Replication factor A protein 1 | | 著者 | Deshpande, I, Seeber, A, Shimada, K, Keusch, J.J, Gut, H, Gasser, S.M. | | 登録日 | 2017-07-28 | | 公開日 | 2017-10-25 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (2.38 Å) | | 主引用文献 | Structural Basis of Mec1-Ddc2-RPA Assembly and Activation on Single-Stranded DNA at Sites of Damage.
Mol. Cell, 68, 2017
|
|
5OMB
 
 | | Crystal structure of K. lactis Ddc2 N-terminus in complex with S. cerevisiae Rfa1 N-OB domain | | 分子名称: | 1,2-ETHANEDIOL, CHLORIDE ION, DNA damage checkpoint protein LCD1, ... | | 著者 | Deshpande, I, Seeber, A, Shimada, K, Keusch, J.J, Gut, H, Gasser, S.M. | | 登録日 | 2017-07-28 | | 公開日 | 2017-10-25 | | 最終更新日 | 2017-11-01 | | 実験手法 | X-RAY DIFFRACTION (1.94 Å) | | 主引用文献 | Structural Basis of Mec1-Ddc2-RPA Assembly and Activation on Single-Stranded DNA at Sites of Damage.
Mol. Cell, 68, 2017
|
|
6FQ3
 
 | | Crystal structure of Danio rerio Lin41 filamin-NHL domains in complex with lin-29A 5'UTR 13mer RNA | | 分子名称: | CHLORIDE ION, E3 ubiquitin-protein ligase TRIM71, RNA (5'-R(*GP*GP*AP*GP*UP*CP*CP*AP*AP*CP*UP*CP*C)-3') | | 著者 | Kumari, P, Aeschimann, F, Gaidatzis, D, Keusch, J.J, Ghosh, P, Neagu, A, Pachulska-Wieczorek, K, Bujnicki, J.M, Gut, H, Grosshans, H, Ciosk, R. | | 登録日 | 2018-02-13 | | 公開日 | 2018-05-09 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (1.901 Å) | | 主引用文献 | Evolutionary plasticity of the NHL domain underlies distinct solutions to RNA recognition.
Nat Commun, 9, 2018
|
|
6FPT
 
 | | Crystal structure of Danio rerio Lin41 filamin-NHL domains | | 分子名称: | E3 ubiquitin-protein ligase TRIM71 | | 著者 | Kumari, P, Aeschimann, F, Gaidatzis, D, Keusch, J.J, Ghosh, P, Neagu, A, Pachulska-Wieczorek, K, Bujnicki, J.M, Gut, H, Grosshans, H, Ciosk, R. | | 登録日 | 2018-02-12 | | 公開日 | 2018-05-09 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Evolutionary plasticity of the NHL domain underlies distinct solutions to RNA recognition.
Nat Commun, 9, 2018
|
|
6FQL
 
 | | Crystal structure of Danio rerio Lin41 filamin-NHL domains in complex with mab-10 3'UTR 13mer RNA | | 分子名称: | CHLORIDE ION, E3 ubiquitin-protein ligase TRIM71, RNA (5'-R(*UP*GP*CP*AP*UP*UP*UP*AP*AP*UP*GP*CP*A)-3') | | 著者 | Kumari, P, Aeschimann, F, Gaidatzis, D, Keusch, J.J, Ghosh, P, Neagu, A, Pachulska-Wieczorek, K, Bujnicki, J.M, Gut, H, Grosshans, H, Ciosk, R. | | 登録日 | 2018-02-14 | | 公開日 | 2018-05-09 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (2.349 Å) | | 主引用文献 | Evolutionary plasticity of the NHL domain underlies distinct solutions to RNA recognition.
Nat Commun, 9, 2018
|
|
