4GTS
 
 | | Engineered RabGGTase in complex with BMS analogue 16 | | Descriptor: | 5-{(3R)-3-(4-hydroxybenzyl)-4-[(4-methoxyphenyl)sulfonyl]-1-[(1-methyl-1H-imidazol-5-yl)methyl]-2,3,4,5-tetrahydro-1H-1,4-benzodiazepin-7-yl}furan-2-carbaldehyde, CALCIUM ION, Geranylgeranyl transferase type-2 subunit alpha, ... | | Authors: | Guo, Z, Stigter, E.A, Bon, R.S, Waldmann, H, Blankenfeldt, W, Goody, R.S. | | Deposit date: | 2012-08-29 | | Release date: | 2012-10-24 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Development of Selective, Potent RabGGTase Inhibitors
J.Med.Chem., 55, 2012
|
|
8Q74
 
 | |
8Q76
 
 | |
8Q75
 
 | |
8Q73
 
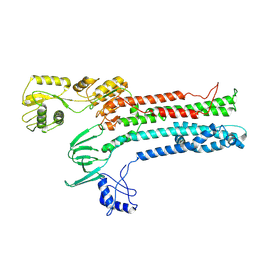 | |
4F38
 
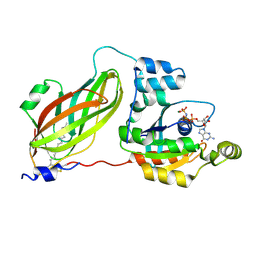 | | Crystal structure of geranylgeranylated RhoA in complex with RhoGDI in its active GPPNHP-bound form | | Descriptor: | GERAN-8-YL GERAN, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, ... | | Authors: | Guo, Z, Tnimov, Z, Alexandrov, K. | | Deposit date: | 2012-05-09 | | Release date: | 2012-05-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Quantitative Analysis of Prenylated RhoA Interaction with Its Chaperone, RhoGDI.
J.Biol.Chem., 287, 2012
|
|
2IGC
 
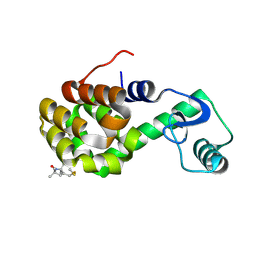 | | Structure of Spin labeled T4 Lysozyme Mutant T115R1A | | Descriptor: | Lysozyme, S-[(1-oxyl-2,2,5,5-tetramethyl-2,5-dihydro-1H-pyrrol-3-yl)methyl] methanesulfonothioate | | Authors: | Guo, Z, Cascio, D, Hideg, K, Hubbell, W.L. | | Deposit date: | 2006-09-22 | | Release date: | 2007-06-12 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural determinants of nitroxide motion in spin-labeled proteins: Tertiary contact and solvent-inaccessible sites in helix G of T4 lysozyme.
Protein Sci., 16, 2007
|
|
7RG7
 
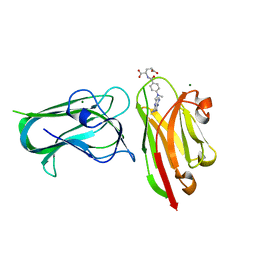 | | Crystal structure of nanoclamp8:VHH in complex with MTX | | Descriptor: | MAGNESIUM ION, METHOTREXATE, nano CLostridial Antibody Mimetic Protein 8 VHH | | Authors: | Guo, Z, Peat, T, Newman, J, Alexandrov, K. | | Deposit date: | 2021-07-14 | | Release date: | 2021-12-22 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Design of a methotrexate-controlled chemical dimerization system and its use in bio-electronic devices.
Nat Commun, 12, 2021
|
|
7RGA
 
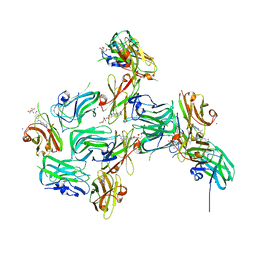 | | Crystal structure of nanoCLAMP3:VHH in complex with MTX | | Descriptor: | METHOTREXATE, SODIUM ION, nano CLostridial Antibody Mimetic Protein 3 VHH | | Authors: | Guo, Z, Alexandrov, K. | | Deposit date: | 2021-07-14 | | Release date: | 2022-05-25 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Design of a methotrexate-controlled chemical dimerization system and its use in bio-electronic devices.
Nat Commun, 12, 2021
|
|
4EHM
 
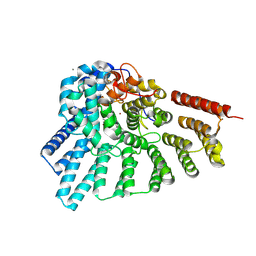 | | RabGGTase in complex with covalently bound Psoromic acid | | Descriptor: | 4-formyl-3-hydroxy-8-methoxy-1,9-dimethyl-11-oxo-11H-dibenzo[b,e][1,4]dioxepine-6-carboxylic acid, CALCIUM ION, Geranylgeranyl transferase type-2 subunit alpha, ... | | Authors: | Guo, Z, Deraeve, C, Bon, R.S, Alexandrov, K, Waldmann, H, Wu, Y.W, Goody, R.S, Blankenfeldt, W. | | Deposit date: | 2012-04-02 | | Release date: | 2012-05-30 | | Last modified: | 2017-08-09 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Psoromic Acid is a Selective and Covalent Rab-Prenylation Inhibitor Targeting Autoinhibited RabGGTase
J.Am.Chem.Soc., 134, 2012
|
|
2OU9
 
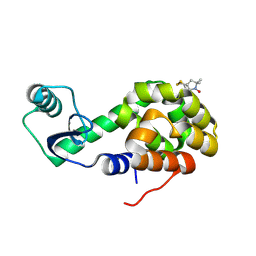 | | Structure of Spin-labeled T4 Lysozyme Mutant T115R1/R119A | | Descriptor: | Lysozyme, S-[(1-oxyl-2,2,5,5-tetramethyl-2,5-dihydro-1H-pyrrol-3-yl)methyl] methanesulfonothioate | | Authors: | Guo, Z, Cascio, D, Hideg, K, Hubbell, W.L. | | Deposit date: | 2007-02-09 | | Release date: | 2007-06-12 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structural determinants of nitroxide motion in spin-labeled proteins: Tertiary contact and solvent-inaccessible sites in helix G of T4 lysozyme.
Protein Sci., 16, 2007
|
|
2NTG
 
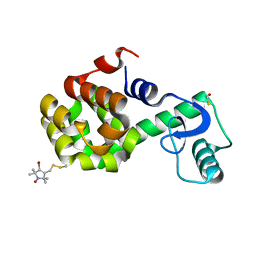 | | Structure of Spin-labeled T4 Lysozyme Mutant T115R7 | | Descriptor: | BETA-MERCAPTOETHANOL, Lysozyme, S-[(4-bromo-1-oxyl-2,2,5,5-tetramethyl-2,5-dihydro-1H-pyrrol-3-yl)methyl] methanesulfonothioate | | Authors: | Guo, Z, Cascio, D, Hideg, K, Hubbell, W.L. | | Deposit date: | 2006-11-07 | | Release date: | 2007-06-12 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural determinants of nitroxide motion in spin-labeled proteins: Tertiary contact and solvent-inaccessible sites in helix G of T4 lysozyme.
Protein Sci., 16, 2007
|
|
2NTH
 
 | | Structure of Spin-labeled T4 Lysozyme Mutant L118R1 | | Descriptor: | Lysozyme, S-[(1-oxyl-2,2,5,5-tetramethyl-2,5-dihydro-1H-pyrrol-3-yl)methyl] methanesulfonothioate | | Authors: | Guo, Z, Cascio, D, Hideg, K, Hubbell, W.L. | | Deposit date: | 2006-11-07 | | Release date: | 2007-06-12 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural determinants of nitroxide motion in spin-labeled proteins: Tertiary contact and solvent-inaccessible sites in helix G of T4 lysozyme.
Protein Sci., 16, 2007
|
|
2Q9D
 
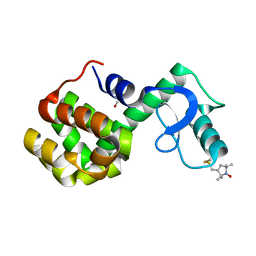 | | Structure of spin-labeled T4 lysozyme mutant A41R1 | | Descriptor: | BETA-MERCAPTOETHANOL, Lysozyme, S-[(1-oxyl-2,2,5,5-tetramethyl-2,5-dihydro-1H-pyrrol-3-yl)methyl] methanesulfonothioate | | Authors: | Guo, Z, Cascio, D, Hideg, K, Hubbell, W.L. | | Deposit date: | 2007-06-12 | | Release date: | 2007-06-26 | | Last modified: | 2023-08-30 | | Method: | EPR (1.4 Å), X-RAY DIFFRACTION | | Cite: | Structural determinants of nitroxide motion in spin-labeled proteins: Solvent-exposed sites in helix B of T4 lysozyme.
Protein Sci., 17, 2008
|
|
2Q9E
 
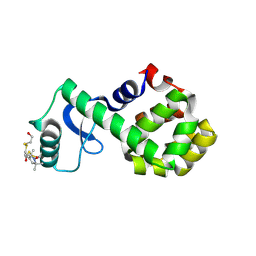 | | Structure of spin-labeled T4 lysozyme mutant S44R1 | | Descriptor: | 2-HYDROXYETHYL DISULFIDE, Lysozyme, S-[(1-oxyl-2,2,5,5-tetramethyl-2,5-dihydro-1H-pyrrol-3-yl)methyl] methanesulfonothioate | | Authors: | Guo, Z, Cascio, D, Hideg, K, Hubbell, W.L. | | Deposit date: | 2007-06-12 | | Release date: | 2007-06-26 | | Last modified: | 2023-08-30 | | Method: | EPR (2.1 Å), X-RAY DIFFRACTION | | Cite: | Structural determinants of nitroxide motion in spin-labeled proteins: Solvent-exposed sites in helix B of T4 lysozyme.
Protein Sci., 17, 2008
|
|
2OU8
 
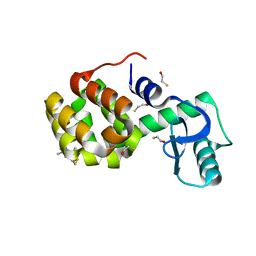 | | Structure of Spin-labeled T4 Lysozyme Mutant T115R1 at Room Temperature | | Descriptor: | BETA-MERCAPTOETHANOL, Lysozyme, S-[(1-oxyl-2,2,5,5-tetramethyl-2,5-dihydro-1H-pyrrol-3-yl)methyl] methanesulfonothioate | | Authors: | Guo, Z, Cascio, D, Hideg, K, Hubbell, W.L. | | Deposit date: | 2007-02-09 | | Release date: | 2007-06-12 | | Last modified: | 2023-08-30 | | Method: | EPR (1.8 Å), X-RAY DIFFRACTION | | Cite: | Structural determinants of nitroxide motion in spin-labeled proteins: Tertiary contact and solvent-inaccessible sites in helix G of T4 lysozyme.
Protein Sci., 16, 2007
|
|
3EUV
 
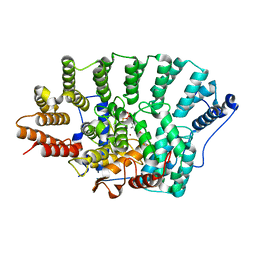 | | Crystal structure of FTase(ALPHA-subunit; BETA-subunit DELTA C10, W102T, Y154T) in complex with BiotinGPP | | Descriptor: | (2E,6E)-3,7-dimethyl-8-({5-[(3aS,4S,6aR)-2-oxohexahydro-1H-thieno[3,4-d]imidazol-4-yl]pentanoyl}amino)octa-2,6-dien-1-yl trihydrogen diphosphate, Protein farnesyltransferase subunit beta, Protein farnesyltransferase/geranylgeranyltransferase type-1 subunit alpha, ... | | Authors: | Guo, Z, Nguyen, U.T.T, Delon, C, Bon, R.S, Blankenfeldt, W, Goody, R.S, Waldmann, H, Wolters, D, Alexandrov, K. | | Deposit date: | 2008-10-11 | | Release date: | 2009-07-07 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Analysis of the eukaryotic prenylome by isoprenoid affinity tagging
Nat.Chem.Biol., 5, 2009
|
|
3EU5
 
 | | Crystal structure of FTase(ALPHA-subunit; BETA-subunit DELTA C10) in complex with BiotinGPP | | Descriptor: | (2E,6E)-3,7-dimethyl-8-({5-[(3aS,4S,6aR)-2-oxohexahydro-1H-thieno[3,4-d]imidazol-4-yl]pentanoyl}amino)octa-2,6-dien-1-yl trihydrogen diphosphate, Protein farnesyltransferase subunit beta, Protein farnesyltransferase/geranylgeranyltransferase type-1 subunit alpha, ... | | Authors: | Guo, Z, Nguyen, U.T.T, Delon, C, Bon, R.S, Blankenfeldt, W, Goody, R.S, Waldmann, H, Wolters, D, Alexandrov, K. | | Deposit date: | 2008-10-09 | | Release date: | 2009-07-07 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Analysis of the eukaryotic prenylome by isoprenoid affinity tagging
Nat.Chem.Biol., 5, 2009
|
|
3DSW
 
 | | Crystal structure of RabGGTase(DELTA LRR; DELTA IG)in complex with mono-prenylated peptide Ser-Cys(GG)-Ser-Cys derivated from Rab7 | | Descriptor: | CALCIUM ION, GERAN-8-YL GERAN, Geranylgeranyl transferase type-2 subunit alpha, ... | | Authors: | Guo, Z, Yu, S, Goody, R.S, Alexandrov, K, Blankenfeldt, W. | | Deposit date: | 2008-07-14 | | Release date: | 2008-09-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structures of RabGGTase-substrate/product complexes provide insights into the evolution of protein prenylation
Embo J., 27, 2008
|
|
3DSU
 
 | | Crystal structure of RabGGTase(DELTA LRR; DELTA IG)in complex with farnesyl pyrophosphate | | Descriptor: | CALCIUM ION, FARNESYL DIPHOSPHATE, Geranylgeranyl transferase type-2 subunit alpha, ... | | Authors: | Guo, Z, Yu, S, Goody, R.S, Alexandrov, K, Blankenfeldt, W. | | Deposit date: | 2008-07-14 | | Release date: | 2008-09-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structures of RabGGTase-substrate/product complexes provide insights into the evolution of protein prenylation
Embo J., 27, 2008
|
|
3DST
 
 | | Crystal structure of RabGGTase(DELTA LRR; DELTA IG)in complex with geranylgeranyl pyrophosphate | | Descriptor: | CALCIUM ION, GERANYLGERANYL DIPHOSPHATE, Geranylgeranyl transferase type-2 subunit alpha, ... | | Authors: | Guo, Z, Yu, S, Goody, R.S, Alexandrov, K, Blankenfeldt, W. | | Deposit date: | 2008-07-14 | | Release date: | 2008-09-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structures of RabGGTase-substrate/product complexes provide insights into the evolution of protein prenylation
Embo J., 27, 2008
|
|
3DSV
 
 | | Crystal structure of RabGGTase(DELTA LRR; DELTA IG)in complex with mono-prenylated peptide Ser-Cys-Ser-Cys(GG) derivated from Rab7 | | Descriptor: | CALCIUM ION, GERAN-8-YL GERAN, Geranylgeranyl transferase type-2 subunit alpha, ... | | Authors: | Guo, Z, Yu, S, Goody, R.S, Alexandrov, K, Blankenfeldt, W. | | Deposit date: | 2008-07-14 | | Release date: | 2008-09-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structures of RabGGTase-substrate/product complexes provide insights into the evolution of protein prenylation
Embo J., 27, 2008
|
|
3PZ4
 
 | | Crystal structure of FTase(ALPHA-subunit; BETA-subunit DELTA C10) in complex with BMS3 and lipid substrate FPP | | Descriptor: | (3R)-3-benzyl-4-[(4-methoxyphenyl)sulfonyl]-1-[(1-methyl-1H-imidazol-5-yl)methyl]-2,3,4,5-tetrahydro-1H-1,4-benzodiazepine-7-carbonitrile, FARNESYL DIPHOSPHATE, Protein farnesyltransferase subunit beta, ... | | Authors: | Guo, Z, Bon, R.S, Stigter, E.A, Waldmann, H, Alexandrov, K, Blankenfeldt, W, Goody, R.S. | | Deposit date: | 2010-12-14 | | Release date: | 2011-05-11 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure-Guided Development of Selective RabGGTase Inhibitors.
Angew.Chem.Int.Ed.Engl., 50, 2011
|
|
3DSX
 
 | | Crystal structure of RabGGTase(DELTA LRR; DELTA IG)in complex with di-prenylated peptide Ser-Cys(GG)-Ser-Cys(GG) derivated from Rab7 | | Descriptor: | CALCIUM ION, GERAN-8-YL GERAN, Geranylgeranyl transferase type-2 subunit alpha, ... | | Authors: | Guo, Z, Yu, S, Goody, R.S, Alexandrov, K, Blankenfeldt, W. | | Deposit date: | 2008-07-14 | | Release date: | 2008-09-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structures of RabGGTase-substrate/product complexes provide insights into the evolution of protein prenylation
Embo J., 27, 2008
|
|
3PZ3
 
 | | Crystal structure of RabGGTase(DELTA LRR; DELTA IG) in Complex with BMS-analogue 14 | | Descriptor: | 4-({(3R)-7-(5-formylfuran-2-yl)-4-[(4-methoxyphenyl)sulfonyl]-1-[(1-methyl-1H-imidazol-5-yl)methyl]-2,3,4,5-tetrahydro-1H-1,4-benzodiazepin-3-yl}methyl)phenyl benzylcarbamate, CALCIUM ION, Geranylgeranyl transferase type-2 subunit alpha, ... | | Authors: | Guo, Z, Bon, R.S, Stigter, E.A, Waldmann, H, Alexandrov, K, Blankenfeldt, W, Goody, R.S. | | Deposit date: | 2010-12-14 | | Release date: | 2011-05-11 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure-Guided Development of Selective RabGGTase Inhibitors.
Angew.Chem.Int.Ed.Engl., 50, 2011
|
|
