4QE9
 
 | | Open MthK pore structure soaked in 10 mM Ba2+/100 mM K+ | | Descriptor: | Calcium-gated potassium channel MthK, POTASSIUM ION | | Authors: | Guo, R, Zeng, W, Cui, H, Chen, L, Ye, S. | | Deposit date: | 2014-05-15 | | Release date: | 2014-07-09 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Ionic interactions of Ba2+ blockades in the MthK K+ channel
J.Gen.Physiol., 144, 2014
|
|
5D8W
 
 | | Structrue of a lucidum protein | | Descriptor: | Endoglucanase | | Authors: | Guo, R, Li, Q, Shang, N, Liu, G, Ko, T.P, Chen, C.C, Liu, W. | | Deposit date: | 2015-08-18 | | Release date: | 2016-06-29 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.86 Å) | | Cite: | Functional and structural analyses of a 1,4-beta-endoglucanase from Ganoderma lucidum.
Enzyme.Microb.Technol., 86, 2016
|
|
5D8Z
 
 | | Structrue of a lucidum protein | | Descriptor: | beta-D-glucopyranose-(1-4)-beta-D-glucopyranose, endoglucanase | | Authors: | Guo, R, Li, Q, Shang, N, Liu, G, Ko, T.P, Chen, C.C, Liu, W. | | Deposit date: | 2015-08-18 | | Release date: | 2016-06-29 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Functional and structural analyses of a 1,4-beta-endoglucanase from Ganoderma lucidum.
Enzyme.Microb.Technol., 86, 2016
|
|
4QE7
 
 | | Open MthK pore structure soaked in 10 mM Ba2+/100 mM Na+ | | Descriptor: | BARIUM ION, Calcium-gated potassium channel MthK | | Authors: | Guo, R, Zeng, W, Cui, H, Chen, L, Ye, S. | | Deposit date: | 2014-05-15 | | Release date: | 2014-07-09 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Ionic interactions of Ba2+ blockades in the MthK K+ channel
J.Gen.Physiol., 144, 2014
|
|
3NBI
 
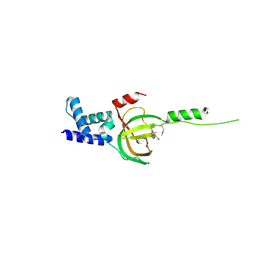 | | Crystal structure of human RMI1 N-terminus | | Descriptor: | RecQ-mediated genome instability protein 1 | | Authors: | Wang, F, Yang, Y, Singh, T.R, Busygina, V, Guo, R, Wan, K, Wang, W, Sung, P, Meetei, A.R, Lei, M. | | Deposit date: | 2010-06-03 | | Release date: | 2010-09-22 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structures of RMI1 and RMI2, Two OB-Fold Regulatory Subunits of the BLM Complex.
Structure, 18, 2010
|
|
3NBH
 
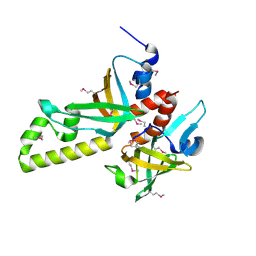 | | Crystal structure of human RMI1C-RMI2 complex | | Descriptor: | RecQ-mediated genome instability protein 1, RecQ-mediated genome instability protein 2 | | Authors: | Wang, F, Yang, Y, Singh, T.R, Busygina, V, Guo, R, Wan, K, Wang, W, Sung, P, Meetei, A.R, Lei, M. | | Deposit date: | 2010-06-03 | | Release date: | 2010-09-22 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structures of RMI1 and RMI2, Two OB-Fold Regulatory Subunits of the BLM Complex.
Structure, 18, 2010
|
|
4WJA
 
 | | Crystal Structure of PAXX | | Descriptor: | Uncharacterized protein C9orf142 | | Authors: | Xing, M, Yang, M, Huo, W, Feng, F, Wei, L, Ning, S, Yan, Z, Li, W, Wang, Q, Hou, M, Dong, C, Guo, R, Gao, G, Ji, J, Lan, L, Liang, H, Xu, D. | | Deposit date: | 2014-09-29 | | Release date: | 2015-03-11 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Interactome analysis identifies a new paralogue of XRCC4 in non-homologous end joining DNA repair pathway.
Nat Commun, 6, 2015
|
|
1NH9
 
 | | Crystal Structure of a DNA Binding Protein Mja10b from the hyperthermophile Methanococcus jannaschii | | Descriptor: | DNA-binding protein Alba | | Authors: | Wang, G, Bartlam, M, Guo, R, Yang, H, Xue, H, Liu, Y, Huang, L, Rao, Z. | | Deposit date: | 2002-12-19 | | Release date: | 2003-12-23 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of a DNA binding protein from the hyperthermophilic euryarchaeon Methanococcus jannaschii
Protein Sci., 12, 2003
|
|
4RO0
 
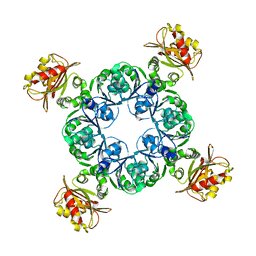 | | Crystal structure of MthK gating ring in a ligand-free form | | Descriptor: | Calcium-gated potassium channel MthK | | Authors: | Dong, W, Guo, R, Chai, H, Chen, Z, Cui, H, Ren, Z, Li, Y, Ye, S. | | Deposit date: | 2014-10-27 | | Release date: | 2015-10-28 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.18 Å) | | Cite: | The principle of cooperative gating in a calcium-gated K+ channel
To be Published
|
|
6YPZ
 
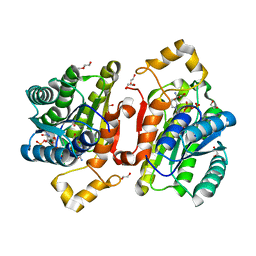 | | Promiscuous Reductase LugOII Catalyzes Keto-reduction at C1 during Lugdunomycin Biosynthesis | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, Monooxygenase, ... | | Authors: | Xiao, X, Elsayed, S.S, Wu, C, van der Heul, H, Prota, A, Huang, J, Guo, R, Abrahams, J.P, van Wezel, G.P. | | Deposit date: | 2020-04-16 | | Release date: | 2020-09-16 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.08 Å) | | Cite: | Functional and Structural Insights into a Novel Promiscuous Ketoreductase of the Lugdunomycin Biosynthetic Pathway.
Acs Chem.Biol., 15, 2020
|
|
6YQ6
 
 | | Promiscuous Reductase LugOII Catalyzes Keto-reduction at C1 during Lugdunomycin Biosynthesis | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, Monooxygenase, ... | | Authors: | Xiao, X, Elsayed, S.S, Wu, C, van der Heul, H, Prota, A, Huang, J, Guo, R, Abrahams, J.P, van Wezel, G.P. | | Deposit date: | 2020-04-16 | | Release date: | 2020-09-16 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Functional and Structural Insights into a Novel Promiscuous Ketoreductase of the Lugdunomycin Biosynthetic Pathway.
Acs Chem.Biol., 15, 2020
|
|
6YQ3
 
 | | Promiscuous Reductase LugOII Catalyzes Keto-reduction at C1 during Lugdunomycin Biosynthesis | | Descriptor: | (3~{R})-8-methoxy-3-methyl-3,6-bis(oxidanyl)-2,4-dihydrobenzo[a]anthracene-1,7,12-trione, 1,2-ETHANEDIOL, Monooxygenase, ... | | Authors: | Xiao, X, Elsayed, S.S, Wu, C, van der Heul, H, Prota, A, Huang, J, Guo, R, Abrahams, J.P, van Wezel, G.P. | | Deposit date: | 2020-04-16 | | Release date: | 2020-09-16 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Functional and Structural Insights into a Novel Promiscuous Ketoreductase of the Lugdunomycin Biosynthetic Pathway.
Acs Chem.Biol., 15, 2020
|
|
6YQ0
 
 | | Promiscuous Reductase LugOII Catalyzes Keto-reduction at C1 during Lugdunomycin Biosynthesis | | Descriptor: | (3~{R})-8-methoxy-3-methyl-3-oxidanyl-2,4-dihydrobenzo[a]anthracene-1,7,12-trione, 1,2-ETHANEDIOL, Monooxygenase, ... | | Authors: | Xiao, X, Elsayed, S.S, Wu, C, van der Heul, H, Prota, A, Huang, J, Guo, R, Abrahams, J.P, van Wezel, G.P. | | Deposit date: | 2020-04-16 | | Release date: | 2020-09-16 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.08 Å) | | Cite: | Functional and Structural Insights into a Novel Promiscuous Ketoreductase of the Lugdunomycin Biosynthetic Pathway.
Acs Chem.Biol., 15, 2020
|
|
4PV7
 
 | | Cocrystal structure of dipeptidyl-peptidase 4 with an indole scaffold inhibitor | | Descriptor: | 1-[2-(2,4-dichlorophenyl)-1-(methylsulfonyl)-1H-indol-3-yl]methanamine, Dipeptidyl peptidase 4 soluble form | | Authors: | Xiao, P, Guo, R, Huang, S, Cui, H, Ye, S, Zhang, Z. | | Deposit date: | 2014-03-15 | | Release date: | 2015-03-18 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.24 Å) | | Cite: | Discovery of dipeptidyl peptidase IV (DPP4) inhibitors based on a novel indole scaffold
Chin.Chem.Lett., 25, 2014
|
|
6M16
 
 | | Cryo-EM structures of SADS-CoV spike glycoproteins | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Wang, X, Yu, J, Qiao, S, Guo, R. | | Deposit date: | 2020-02-24 | | Release date: | 2020-05-27 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (2.83 Å) | | Cite: | Cryo-EM structures of HKU2 and SADS-CoV spike glycoproteins provide insights into coronavirus evolution.
Nat Commun, 11, 2020
|
|
6M15
 
 | | Cryo-EM structures of HKU2 spike glycoproteins | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Wang, X, Yu, J, Qiao, S, Guo, R. | | Deposit date: | 2020-02-24 | | Release date: | 2020-05-27 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (2.38 Å) | | Cite: | Cryo-EM structures of HKU2 and SADS-CoV spike glycoproteins provide insights into coronavirus evolution.
Nat Commun, 11, 2020
|
|
5GPN
 
 | | Architecture of mammalian respirasome | | Descriptor: | Acyl carrier protein, COPPER (II) ION, Cytochrome b, ... | | Authors: | Gu, J, Wu, M, Guo, R, Yang, M. | | Deposit date: | 2016-08-03 | | Release date: | 2017-04-05 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (5.4 Å) | | Cite: | The architecture of the mammalian respirasome.
Nature, 537, 2016
|
|
5GUP
 
 | | Cryo-EM structure of mammalian respiratory supercomplex I1III2IV1 | | Descriptor: | (9R,11S)-9-({[(1S)-1-HYDROXYHEXADECYL]OXY}METHYL)-2,2-DIMETHYL-5,7,10-TRIOXA-2LAMBDA~5~-AZA-6LAMBDA~5~-PHOSPHAOCTACOSANE-6,6,11-TRIOL, 1,2-Dioleoyl-sn-glycero-3-phosphoethanolamine, Acyl carrier protein, ... | | Authors: | Gu, J, Wu, M, Guo, R, Yang, M. | | Deposit date: | 2016-08-30 | | Release date: | 2017-03-29 | | Last modified: | 2019-11-06 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structure of Mammalian Respiratory Supercomplex I1III2IV1
Cell, 167, 2016
|
|
7CWH
 
 | | Structural basis of RACK7 PHD to read a pediatric glioblastoma-associated histone mutation H3.3G34R | | Descriptor: | Peptide from Histone H3.3, Protein kinase C-binding protein 1, ZINC ION | | Authors: | Lan, W.X, Li, Z, Jiao, F.F, Wang, C.X, Guo, R, Cao, C.Y. | | Deposit date: | 2020-08-28 | | Release date: | 2021-05-26 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural basis of RACK7 PHD domain to read a pediatric glioblastoma‐associated histone mutation H3.3G34R
Chin.J.Chem., 2021
|
|
7CWZ
 
 | | Crystal structure of a tyrosine decarboxylase from Enterococcus faecalis K392A mutant in complex with the cofactor PLP and L-dopa | | Descriptor: | Decarboxylase, L-DOPAMINE, MAGNESIUM ION, ... | | Authors: | Yu, X, Gong, M, Huang, J, Liu, W, Chen, C, Guo, R. | | Deposit date: | 2020-09-01 | | Release date: | 2021-09-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.97 Å) | | Cite: | Crystal structure of a tyrosine decarboxylase from Enterococcus faecalis K392A mutant in complex with the cofactor PLP and L-dopa
to be published
|
|
7CX0
 
 | | Crystal structure of a tyrosine decarboxylase from Enterococcus faecalis in complex with the cofactor PLP and inhibitor carbidopa | | Descriptor: | CARBIDOPA, Decarboxylase, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Yu, X, Gong, M, Huang, J, Liu, W, Chen, C, Guo, R. | | Deposit date: | 2020-09-01 | | Release date: | 2021-09-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.66 Å) | | Cite: | Crystal structure of a tyrosine decarboxylase from Enterococcus faecalis in complex with the cofactor PLP and inhibitor carbidopa
to be published
|
|
7CWY
 
 | | Crystal structure of a tyrosine decarboxylase from Enterococcus faecalis in complex with the cofactor PLP | | Descriptor: | Decarboxylase | | Authors: | Yu, X, Gong, M, Huang, J, Liu, W, Chen, C, Guo, R. | | Deposit date: | 2020-09-01 | | Release date: | 2021-09-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Crystal structure of a tyrosine decarboxylase from Enterococcus faecalis in complex with the cofactor PLP
to be published
|
|
7CWX
 
 | | Crystal structure of a tyrosine decarboxylase from Enterococcus faecalis | | Descriptor: | DI(HYDROXYETHYL)ETHER, Decarboxylase, GLYCEROL | | Authors: | Yu, X, Gong, M, Huang, J, Liu, W, Chen, C, Guo, R. | | Deposit date: | 2020-09-01 | | Release date: | 2021-09-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal structure of a tyrosine decarboxylase from Enterococcus faecalis
to be published
|
|
7CX1
 
 | | Crystal structure of a tyrosine decarboxylase from Enterococcus faecalis in complex with the cofactor PLP and inhibitor methyl-tyrosine | | Descriptor: | 4-[(2R)-2-(methylamino)propyl]phenol, Decarboxylase | | Authors: | Yu, X, Gong, M, Huang, J, Liu, W, Chen, C, Guo, R. | | Deposit date: | 2020-09-01 | | Release date: | 2021-09-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.54 Å) | | Cite: | Crystal structure of a tyrosine decarboxylase from Enterococcus faecalis in complex with the cofactor PLP and inhibitor methyl-tyrosine
to be published
|
|
7W5G
 
 | | The apo structure of trichobrasilenol synthase TaTC6 with the space group of orthorhombic | | Descriptor: | GLYCEROL, SULFATE ION, Terpene cyclase 6 | | Authors: | Chen, C, Wang, T, Yang, Y, Zhang, L, Ko, T, Huang, J, Guo, R. | | Deposit date: | 2021-11-30 | | Release date: | 2022-06-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural insights into the cyclization of unusual brasilane-type sesquiterpenes.
Int.J.Biol.Macromol., 209, 2022
|
|
