5GT7
 
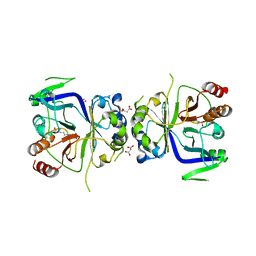 | | Crystal Structure of Arg-bound CASTOR1 | | Descriptor: | ARGININE, GATS-like protein 3, MALONATE ION | | Authors: | Guo, L, Deng, D. | | Deposit date: | 2016-08-18 | | Release date: | 2017-08-23 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.048 Å) | | Cite: | Crystal structures of arginine sensor CASTOR1 in arginine-bound and ligand free states
Biochem. Biophys. Res. Commun., 508, 2019
|
|
6JBT
 
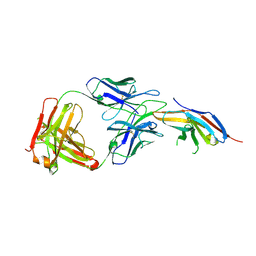 | | Complex structure of toripalimab-Fab and PD-1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain, ... | | Authors: | Guo, L, Tan, S, Chai, Y, Qi, J, Gao, G.F, Yan, J. | | Deposit date: | 2019-01-26 | | Release date: | 2019-06-19 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.47 Å) | | Cite: | Glycosylation-independent binding of monoclonal antibody toripalimab to FG loop of PD-1 for tumor immune checkpoint therapy.
Mabs, 11, 2019
|
|
2H8N
 
 | |
2O94
 
 | |
5GT8
 
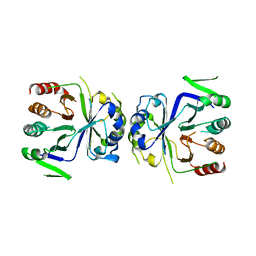 | | Crystal Structure of apo-CASTOR1 | | Descriptor: | GATS-like protein 3 | | Authors: | Guo, L, Deng, D. | | Deposit date: | 2016-08-18 | | Release date: | 2017-08-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structures of arginine sensor CASTOR1 in arginine-bound and ligand free states
Biochem. Biophys. Res. Commun., 508, 2019
|
|
6J6V
 
 | |
5YY8
 
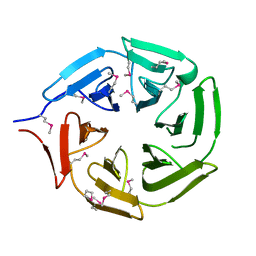 | | Crystal structure of the Kelch domain of human NS1-BP | | Descriptor: | Influenza virus NS1A-binding protein | | Authors: | Guo, L, Liu, Y, Liang, H. | | Deposit date: | 2017-12-08 | | Release date: | 2018-03-14 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.979 Å) | | Cite: | Crystal structure of the Kelch domain of human NS1-binding protein at 1.98 angstrom resolution.
Acta Crystallogr F Struct Biol Commun, 74, 2018
|
|
7CT5
 
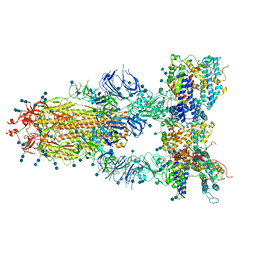 | | S protein of SARS-CoV-2 in complex bound with T-ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, ... | | Authors: | Guo, L, Bi, W.W, Zhang, Y.Y, Yan, R.H, Li, Y.N, Zhou, Q, Dang, B.B. | | Deposit date: | 2020-08-18 | | Release date: | 2020-11-18 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Engineered trimeric ACE2 binds viral spike protein and locks it in "Three-up" conformation to potently inhibit SARS-CoV-2 infection.
Cell Res., 31, 2021
|
|
8Y4H
 
 | |
8Z4F
 
 | |
3LWI
 
 | | Crystal structure of Cren7-dsDNA complex | | Descriptor: | Chromatin protein Cren7, DNA (5'-D(*GP*CP*GP*AP*TP*CP*GP*C)-3') | | Authors: | Zhang, Z.F, Gong, Y, Guo, L, Jiang, T, Huang, L. | | Deposit date: | 2010-02-23 | | Release date: | 2010-05-26 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural insights into the interaction of the crenarchaeal chromatin protein Cren7 with DNA
Mol.Microbiol., 76, 2010
|
|
3LWH
 
 | | Crystal structure of Cren7-dsDNA complex | | Descriptor: | Chromatin protein Cren7, DNA (5'-D(*GP*TP*AP*AP*TP*TP*AP*C)-3') | | Authors: | Zhang, Z.F, Gong, Y, Guo, L, Jiang, T, Huang, L. | | Deposit date: | 2010-02-23 | | Release date: | 2010-05-26 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural insights into the interaction of the crenarchaeal chromatin protein Cren7 with DNA
Mol.Microbiol., 76, 2010
|
|
9JA8
 
 | | Crystal structure of human phosphodiesterase 10A in complex with N-(2-amino-2-thioxoethyl)-2-(3-(3-(dimethylcarbamoyl)-6-fluoroimidazo[1,2-a]pyridin-2-yl)azetidin-1-yl)quinoline-4-carboxamide | | Descriptor: | MAGNESIUM ION, ZINC ION, cAMP and cAMP-inhibited cGMP 3',5'-cyclic phosphodiesterase 10A, ... | | Authors: | Zhang, F.C, Huang, Y.Y, Luo, H.B, Guo, L. | | Deposit date: | 2024-08-24 | | Release date: | 2025-01-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Inhalable Carbonyl Sulfide Donor-Hybridized Selective Phosphodiesterase 10A Inhibitor for Treating Idiopathic Pulmonary Fibrosis by Inhibiting Tumor Growth Factor-beta Signaling and Activating the cAMP/Protein Kinase A/cAMP Response Element-Binding Protein (CREB)/p53 Axis.
Acs Pharmacol Transl Sci, 8, 2025
|
|
6JIF
 
 | | Crystal Structures of Branched-Chain Aminotransferase from Pseudomonas sp. UW4 | | Descriptor: | Branched-chain-amino-acid aminotransferase, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Zheng, X, Guo, L, Li, D.F, Wu, B. | | Deposit date: | 2019-02-21 | | Release date: | 2020-01-01 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Biochemical and structural characterization of a highly active branched-chain amino acid aminotransferase from Pseudomonas sp. for efficient biosynthesis of chiral amino acids.
Appl.Microbiol.Biotechnol., 103, 2019
|
|
7XUZ
 
 | | Crystal structure of a HDAC4-MEF2A-DNA ternary complex | | Descriptor: | DNA (5'-D(*TP*CP*TP*TP*AP*TP*AP*AP*AP*TP*AP*GP*T)-3'), DNA (5'-D(P*AP*CP*TP*AP*TP*TP*TP*AP*TP*AP*A)-3'), Histone deacetylase 4, ... | | Authors: | Dai, S.Y, Guo, L, Dey, R, Guo, M, Bates, D, Cayford, J, Chen, X.J, Wei, X.D, Chen, L, Chen, Y.H. | | Deposit date: | 2022-05-20 | | Release date: | 2023-11-29 | | Last modified: | 2024-06-12 | | Method: | X-RAY DIFFRACTION (3.591 Å) | | Cite: | Structural insights into the HDAC4-MEF2A-DNA complex and its implication in long-range transcriptional regulation.
Nucleic Acids Res., 52, 2024
|
|
2JTM
 
 | | Solution structure of Sso6901 from Sulfolobus solfataricus P2 | | Descriptor: | Putative uncharacterized protein | | Authors: | Feng, Y, Guo, L, Huang, L, Wang, J. | | Deposit date: | 2007-08-03 | | Release date: | 2008-04-29 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Biochemical and structural characterization of Cren7, a novel chromatin protein conserved among Crenarchaea
Nucleic Acids Res., 36, 2008
|
|
3TLQ
 
 | | Crystal structure of EAL-like domain protein YdiV | | Descriptor: | GLYCEROL, PHOSPHATE ION, Regulatory protein YdiV | | Authors: | Li, B, Li, N, Wang, F, Guo, L, Liu, C, Zhu, D, Xu, S, Gu, L. | | Deposit date: | 2011-08-30 | | Release date: | 2012-09-12 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Structural insight of a concentration-dependent mechanism by which YdiV inhibits Escherichia coli flagellum biogenesis and motility
Nucleic Acids Res., 40, 2012
|
|
2AS5
 
 | | Structure of the DNA binding domains of NFAT and FOXP2 bound specifically to DNA. | | Descriptor: | 5'-D(AP*AP*CP*TP*AP*TP*GP*AP*AP*AP*CP*AP*AP*AP*TP*TP*TP*TP*CP*CP*TP*)-3', 5'-D(TP*TP*AP*GP*GP*AP*AP*AP*AP*TP*TP*TP*GP*TP*TP*TP*CP*AP*TP*AP*GP*)-3', Forkhead box protein P2, ... | | Authors: | Wu, Y, Stroud, J.C, Borde, M, Bates, D.L, Guo, L, Han, A, Rao, A, Chen, L. | | Deposit date: | 2005-08-22 | | Release date: | 2006-08-08 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | FOXP3 Controls Regulatory T Cell Function through Cooperation with NFAT.
Cell(Cambridge,Mass.), 126, 2006
|
|
2DR2
 
 | | Structure of human tryptophanyl-tRNA synthetase in complex with tRNA(Trp) | | Descriptor: | SULFATE ION, TRYPTOPHAN, Tryptophanyl-tRNA synthetase, ... | | Authors: | Shen, N, Guo, L, Yang, B, Jin, Y, Ding, J. | | Deposit date: | 2006-06-05 | | Release date: | 2006-07-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure of human tryptophanyl-tRNA synthetase in complex with tRNA(Trp) reveals the molecular basis of tRNA recognition and specificity
Nucleic Acids Res., 34, 2006
|
|
3DFX
 
 | | Opposite GATA DNA binding | | Descriptor: | DNA (5'-D(*DAP*DAP*DGP*DGP*DTP*DTP*DAP*DTP*DCP*DTP*DCP*DTP*DGP*DAP*DTP*DTP*DTP*DAP*DTP*DC)-3'), DNA (5'-D(*DTP*DTP*DGP*DAP*DTP*DAP*DAP*DAP*DTP*DCP*DAP*DGP*DAP*DGP*DAP*DTP*DAP*DAP*DCP*DC)-3'), Trans-acting T-cell-specific transcription factor GATA-3, ... | | Authors: | Bates, D.L, Kim, G.K, Guo, L, Chen, L. | | Deposit date: | 2008-06-12 | | Release date: | 2008-07-29 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structures of multiple GATA zinc fingers bound to DNA reveal new insights into DNA recognition and self-association by GATA.
J.Mol.Biol., 381, 2008
|
|
3ODO
 
 | |
3ODW
 
 | |
3P6A
 
 | |
3M0E
 
 | | Crystal structure of the ATP-bound state of Walker B mutant of NtrC1 ATPase domain | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Transcriptional regulator (NtrC family) | | Authors: | Chen, B, Sysoeva, T.A, Chowdhury, S, Rusu, M, Birmanns, S, Guo, L, Hanson, J, Yang, H, Nixon, B.T. | | Deposit date: | 2010-03-02 | | Release date: | 2010-11-03 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.63 Å) | | Cite: | Engagement of Arginine Finger to ATP Triggers Large Conformational Changes in NtrC1 AAA+ ATPase for Remodeling Bacterial RNA Polymerase.
Structure, 18, 2010
|
|
3ODX
 
 | |
