4CG4
 
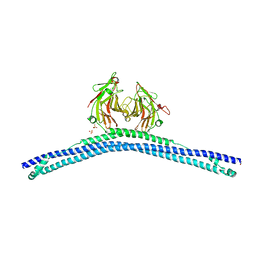 | | Crystal structure of the CHS-B30.2 domains of TRIM20 | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-ETHANETHIOL, PYRIN, ... | | Authors: | Weinert, C, Morger, D, Djekic, A, Mittl, P.R.E, Gruetter, M.G. | | Deposit date: | 2013-11-20 | | Release date: | 2015-04-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structure of Trim20 C-Terminal Coiled-Coil/B30.2 Fragment: Implications for the Recognition of Higher Order Oligomers
Sci.Rep., 5, 2015
|
|
1HII
 
 | |
1HIH
 
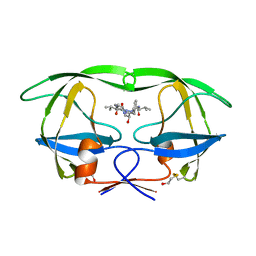 | |
4BS0
 
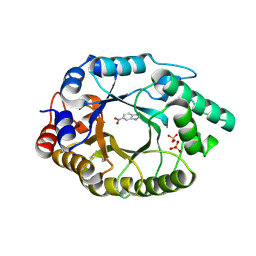 | | Crystal Structure of Kemp Eliminase HG3.17 E47N,N300D Complexed with Transition State Analog 6-Nitrobenzotriazole | | Descriptor: | 6-NITROBENZOTRIAZOLE, KEMP ELIMINASE HG3.17, SULFATE ION | | Authors: | Blomberg, R, Kries, H, Pinkas, D.M, Mittl, P.R.E, Gruetter, M.G, Privett, H.K, Mayo, S, Hilvert, D. | | Deposit date: | 2013-06-06 | | Release date: | 2013-10-16 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.09 Å) | | Cite: | Precision is Essential for Efficient Catalysis in an Evolved Kemp Eliminase
Nature, 503, 2013
|
|
4HJ0
 
 | | Crystal structure of the human GIPr ECD in complex with Gipg013 Fab at 3-A resolution | | Descriptor: | Gastric inhibitory polypeptide receptor, Gipg013 Fab, Antagonizing antibody to the GIP Receptor, ... | | Authors: | Madhurantakam, C, Ravn, P, Gruetter, M.G, Jackson, R.H. | | Deposit date: | 2012-10-12 | | Release date: | 2013-05-29 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural and Pharmacological Characterization of Novel Potent and Selective Monoclonal Antibody Antagonists of Glucose-dependent Insulinotropic Polypeptide Receptor.
J.Biol.Chem., 288, 2013
|
|
4J7W
 
 | | E3_5 DARPin L86A mutant | | Descriptor: | DARPin_E3_5_L86A | | Authors: | Seeger, M.A, Gruetter, M.G. | | Deposit date: | 2013-02-14 | | Release date: | 2013-08-14 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Design, construction, and characterization of a second-generation DARPin library with reduced hydrophobicity.
Protein Sci., 22, 2013
|
|
4J8Y
 
 | | E3_5 DARPin D77S mutant | | Descriptor: | DARPin_E3_5_D77S | | Authors: | Seeger, M.A, Gruetter, M.G. | | Deposit date: | 2013-02-15 | | Release date: | 2013-08-14 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Design, construction, and characterization of a second-generation DARPin library with reduced hydrophobicity.
Protein Sci., 22, 2013
|
|
4JB8
 
 | | Caspase-7 in Complex with DARPin C7_16 | | Descriptor: | Caspase-7 subunit p11, Caspase-7 subunit p20, DARPin C7_16 | | Authors: | Fluetsch, A, Seeger, M.A, Gruetter, M.G. | | Deposit date: | 2013-02-19 | | Release date: | 2013-08-21 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Design, construction, and characterization of a second-generation DARPin library with reduced hydrophobicity.
Protein Sci., 22, 2013
|
|
4I2O
 
 | | The Structure of FixK2 from Bradyrhizobium japonicum | | Descriptor: | FixK2 protein, Promoter of fixK2 direct target, fixN, ... | | Authors: | Bonnet, M, Kurz, M, Mesa, S, Briand, C, Hennecke, H, Gruetter, M.G. | | Deposit date: | 2012-11-22 | | Release date: | 2013-04-10 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | The Structure of Bradyrhizobium japonicum Transcription Factor FixK2 Unveils Sites of DNA Binding and Oxidation.
J.Biol.Chem., 288, 2013
|
|
2V7N
 
 | | Unusual twinning in crystals of the CitS binding antibody Fab fragment f3p4 | | Descriptor: | IMMUNOGLOBULIN HEAVY CHAIN, IMMUNOGLOBULIN LIGHT CHAIN | | Authors: | Frey, D, Huber, T, Plueckthun, A, Gruetter, M.G. | | Deposit date: | 2007-07-31 | | Release date: | 2008-06-17 | | Last modified: | 2019-05-22 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Structure of the Recombinant Antibody Fab Fragment F3P4.
Acta Crystallogr.,Sect.D, 64, 2008
|
|
2V50
 
 | |
2AXI
 
 | | HDM2 in complex with a beta-hairpin | | Descriptor: | 3[N-MORPHOLINO]PROPANE SULFONIC ACID, SULFATE ION, Ubiquitin-protein ligase E3 Mdm2, ... | | Authors: | Mittl, P.R.E, Fasan, R, Robinson, J, Gruetter, M.G. | | Deposit date: | 2005-09-05 | | Release date: | 2006-03-21 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structure-Activity Studies in a Family of beta-Hairpin Protein Epitope Mimetic Inhibitors of the p53-HDM2 Protein-Protein Interaction.
Chembiochem, 7, 2006
|
|
4DX5
 
 | | Transport of drugs by the multidrug transporter AcrB involves an access and a deep binding pocket that are separated by a switch-loop | | Descriptor: | (4S,4AS,5AR,12AS)-4,7-BIS(DIMETHYLAMINO)-3,10,12,12A-TETRAHYDROXY-1,11-DIOXO-1,4,4A,5,5A,6,11,12A-OCTAHYDROTETRACENE-2- CARBOXAMIDE, Acriflavine resistance protein B, DARPIN, ... | | Authors: | Eicher, T, Cha, H, Seeger, M.A, Brandstaetter, L, El-Delik, J, Bohnert, J.A, Kern, W.V, Verrey, F, Gruetter, M.G, Diederichs, K, Pos, K.M. | | Deposit date: | 2012-02-27 | | Release date: | 2012-05-02 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Transport of drugs by the multidrug transporter AcrB involves an access and a deep binding pocket that are separated by a switch-loop.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4DX7
 
 | | Transport of drugs by the multidrug transporter AcrB involves an access and a deep binding pocket that are separated by a switch-loop | | Descriptor: | Acriflavine resistance protein B, DARPIN, DECANE, ... | | Authors: | Eicher, T, Cha, H, Seeger, M.A, Brandstaetter, L, El-Delik, J, Bohnert, J.A, Kern, W.V, Verrey, F, Gruetter, M.G, Diederichs, K, Pos, K.M. | | Deposit date: | 2012-02-27 | | Release date: | 2012-05-02 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.253 Å) | | Cite: | Transport of drugs by the multidrug transporter AcrB involves an access and a deep binding pocket that are separated by a switch-loop.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4DX6
 
 | | Transport of drugs by the multidrug transporter AcrB involves an access and a deep binding pocket that are separated by a switch-loop | | Descriptor: | Acriflavine resistance protein B, DARPIN, DODECYL-BETA-D-MALTOSIDE | | Authors: | Eicher, T, Cha, H, Seeger, M.A, Brandstaetter, L, El-Delik, J, Bohnert, J.A, Kern, W.V, Verrey, F, Gruetter, M.G, Diederichs, K, Pos, K.M. | | Deposit date: | 2012-02-27 | | Release date: | 2012-05-02 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Transport of drugs by the multidrug transporter AcrB involves an access and a deep binding pocket that are separated by a switch-loop.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
1AIE
 
 | |
3NFB
 
 | | Crystal structure of the N-terminal beta-aminopeptidase BapA in complex with hydrolyzed ampicillin | | Descriptor: | (2S,4S)-2-[(R)-{[(2R)-2-amino-2-phenylacetyl]amino}(carboxy)methyl]-5,5-dimethyl-1,3-thiazolidine-4-carboxylic acid, Beta-peptidyl aminopeptidase, GLYCEROL, ... | | Authors: | Merz, T, Heck, T, Geueke, B, Kohler, H.-P.E, Gruetter, M.G. | | Deposit date: | 2010-06-10 | | Release date: | 2011-09-28 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure and inhibition of the beta-aminopeptidase BapA, a new ampicillin-recognizing member of the N-terminal nucleophile hydrolase family
To be Published
|
|
2P2C
 
 | | Inhibition of caspase-2 by a designed ankyrin repeat protein (DARPin) | | Descriptor: | Caspase-2 | | Authors: | Roschitzki Voser, H, Briand, C, Capitani, G, Gruetter, M.G. | | Deposit date: | 2007-03-07 | | Release date: | 2007-05-22 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.24 Å) | | Cite: | Inhibition of Caspase-2 by a Designed Ankyrin Repeat Protein: Specificity, Structure, and Inhibition Mechanism.
Structure, 15, 2007
|
|
3NDV
 
 | | Crystal structure of the N-terminal beta-aminopeptidase BapA in complex with ampicillin | | Descriptor: | (2S,4S)-2-[(1R)-1-{[(2R)-2-amino-2-phenylacetyl]amino}-2-oxoethyl]-5,5-dimethyl-1,3-thiazolidine-4-carboxylic acid, (2S,5R,6R)-6-{[(2R)-2-AMINO-2-PHENYLETHANOYL]AMINO}-3,3-DIMETHYL-7-OXO-4-THIA-1-AZABICYCLO[3.2.0]HEPTANE-2-CARBOXYLIC ACID, Beta-peptidyl aminopeptidase, ... | | Authors: | Merz, T, Heck, T, Geueke, B, Kohler, H.-P.E, Gruetter, M.G. | | Deposit date: | 2010-06-08 | | Release date: | 2011-09-28 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structures and inhibition of the beta-aminopeptidase BapA, a new ampicillin-recognizing member of the N-terminal nucleophile hydrolase family
To be Published
|
|
2FWG
 
 | | high resolution crystal structure of the C-terminal domain of the electron transfer catalyst DsbD (photoreduced form) | | Descriptor: | Thiol:disulfide interchange protein dsbD | | Authors: | Stirnimann, C.U, Rozhkova, A, Grauschopf, U, Boeckmann, R.A, Glockshuber, R, Capitani, G, Gruetter, M.G. | | Deposit date: | 2006-02-02 | | Release date: | 2006-06-13 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | High-resolution structures of Escherichia coli cDsbD in different redox states: A combined crystallographic, biochemical and computational study
J.Mol.Biol., 358, 2006
|
|
2FWH
 
 | | atomic resolution crystal structure of the C-terminal domain of the electron transfer catalyst DsbD (reduced form at pH7) | | Descriptor: | DI(HYDROXYETHYL)ETHER, IODIDE ION, Thiol:disulfide interchange protein dsbD | | Authors: | Stirnimann, C.U, Rozhkova, A, Grauschopf, U, Boeckmann, R.A, Glockshuber, R, Capitani, G, Gruetter, M.G. | | Deposit date: | 2006-02-02 | | Release date: | 2006-06-13 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (0.99 Å) | | Cite: | High-resolution structures of Escherichia coli cDsbD in different redox states: A combined crystallographic, biochemical and computational study
J.Mol.Biol., 358, 2006
|
|
2FWF
 
 | | high resolution crystal structure of the C-terminal domain of the electron transfer catalyst DsbD (reduced form) | | Descriptor: | IODIDE ION, SODIUM ION, Thiol:disulfide interchange protein dsbD | | Authors: | Stirnimann, C.U, Rozhkova, A, Grauschopf, U, Boeckmann, R.A, Glockshuber, R, Capitani, G, Gruetter, M.G. | | Deposit date: | 2006-02-02 | | Release date: | 2006-06-13 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | High-resolution structures of Escherichia coli cDsbD in different redox states: A combined crystallographic, biochemical and computational study
J.Mol.Biol., 358, 2006
|
|
1BX7
 
 | | HIRUSTASIN FROM HIRUDO MEDICINALIS AT 1.2 ANGSTROMS | | Descriptor: | HIRUSTASIN, SULFATE ION | | Authors: | Uson, I, Sheldrick, G.M, De La Fortelle, E, Bricogne, G, Di Marco, S, Priestle, J.P, Gruetter, M.G, Mittl, P.R.E. | | Deposit date: | 1998-10-14 | | Release date: | 1999-04-27 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | The 1.2 A crystal structure of hirustasin reveals the intrinsic flexibility of a family of highly disulphide-bridged inhibitors.
Structure Fold.Des., 7, 1999
|
|
1BX8
 
 | | HIRUSTASIN FROM HIRUDO MEDICINALIS AT 1.4 ANGSTROMS | | Descriptor: | HIRUSTASIN, SULFATE ION | | Authors: | Uson, I, Sheldrick, G.M, De La Fortelle, E, Bricogne, G, Di Marco, S, Priestle, J.P, Gruetter, M.G, Mittl, P.R.E. | | Deposit date: | 1998-10-14 | | Release date: | 1999-04-27 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | The 1.2 A crystal structure of hirustasin reveals the intrinsic flexibility of a family of highly disulphide-bridged inhibitors.
Structure Fold.Des., 7, 1999
|
|
1TMT
 
 | |
