4FQF
 
 | |
4FR8
 
 | | Crystal structure of human aldehyde dehydrogenase-2 in complex with nitroglycerin | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ADENOSINE-5'-DIPHOSPHATE, Aldehyde dehydrogenase, ... | | Authors: | Lang, B.S, Gruber, K. | | Deposit date: | 2012-06-26 | | Release date: | 2012-09-26 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Vascular Bioactivation of Nitroglycerin by Aldehyde Dehydrogenase-2: REACTION INTERMEDIATES REVEALED BY CRYSTALLOGRAPHY AND MASS SPECTROMETRY.
J.Biol.Chem., 287, 2012
|
|
3FWA
 
 | |
3FW7
 
 | |
3FW8
 
 | |
4FQD
 
 | |
5L46
 
 | |
5LUI
 
 | | Structure of cutinase 1 from Thermobifida cellulosilytica | | Descriptor: | CHLORIDE ION, Cutinase 1, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Hromic, A, Lyskowski, A, Gruber, K. | | Deposit date: | 2016-09-08 | | Release date: | 2017-07-12 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Small cause, large effect: Structural characterization of cutinases from Thermobifida cellulosilytica.
Biotechnol. Bioeng., 114, 2017
|
|
4EC3
 
 | | Structure of berberine bridge enzyme, H174A variant in complex with (S)-reticuline | | Descriptor: | (S)-reticuline, 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, ... | | Authors: | Winkler, A, Macheroux, P, Gruber, K. | | Deposit date: | 2012-03-26 | | Release date: | 2012-07-18 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.6501 Å) | | Cite: | Catalytic and structural role of a conserved active site histidine in berberine bridge enzyme.
Biochemistry, 51, 2012
|
|
1ZCH
 
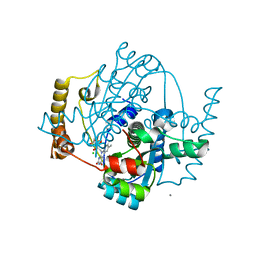 | | Structure of the hypothetical oxidoreductase YcnD from Bacillus subtilis | | Descriptor: | CALCIUM ION, CHLORIDE ION, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Morokutti, A, Lyskowski, A, Sollner, S, Pointner, E, Fitzpatrick, T.B, Kratky, C, Gruber, K, Macheroux, P. | | Deposit date: | 2005-04-12 | | Release date: | 2005-11-01 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure and Function of YcnD from Bacillus subtilis, a Flavin-Containing Oxidoreductase(,).
Biochemistry, 44, 2005
|
|
3T6B
 
 | |
5MP4
 
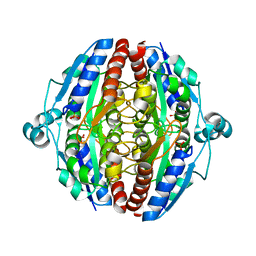 | | The structure of Pst2p from Saccharomyces cerevisiae | | Descriptor: | PHOSPHATE ION, Protoplast secreted protein 2 | | Authors: | Hromic, A, Gruber, K. | | Deposit date: | 2016-12-15 | | Release date: | 2017-05-24 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | Structure, biochemical and kinetic properties of recombinant Pst2p from Saccharomyces cerevisiae, a FMN-dependent NAD(P)H:quinone oxidoreductase.
Biochim. Biophys. Acta, 1865, 2017
|
|
3T6J
 
 | |
8ACS
 
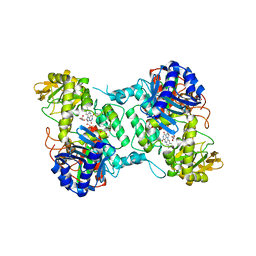 | | Crystal structure of FMO from Janthinobacterium svalbardensis | | Descriptor: | DI(HYDROXYETHYL)ETHER, FAD-dependent oxidoreductase, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Polidori, N, Galuska, P, Gruber, K. | | Deposit date: | 2022-07-06 | | Release date: | 2022-09-07 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A Cold-Active Flavin-Dependent Monooxygenase from Janthinobacterium svalbardensis Unlocks Applications of Baeyer-Villiger Monooxygenases at Low Temperature.
Acs Catalysis, 13, 2023
|
|
5NY5
 
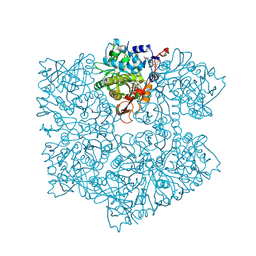 | | The apo structure of 3,4-dihydroxybenzoic acid decarboxylases from Enterobacter cloacae | | Descriptor: | 3,4-dihydroxybenzoate decarboxylase, GLYCEROL | | Authors: | Dordic, A, Gruber, K, Payer, S, Glueck, S, Pavkov-Keller, T, Marshall, S, Leys, D. | | Deposit date: | 2017-05-11 | | Release date: | 2017-09-13 | | Last modified: | 2020-11-18 | | Method: | X-RAY DIFFRACTION (2.501 Å) | | Cite: | Regioselective para-Carboxylation of Catechols with a Prenylated Flavin Dependent Decarboxylase.
Angew. Chem. Int. Ed. Engl., 56, 2017
|
|
8AUG
 
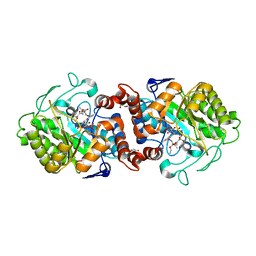 | |
8AUM
 
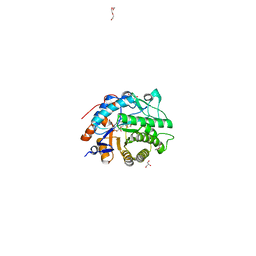 | |
8AUN
 
 | |
8AUE
 
 | |
8AU9
 
 | |
8AUB
 
 | |
8A8I
 
 | | Xenobiotic reductase A from Pseudomonas putida in complex with ethyl (Z)-2-(hydroxyimino)-3-oxobutanoate | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, ... | | Authors: | Polidori, N, Gruber, K. | | Deposit date: | 2022-06-23 | | Release date: | 2023-03-01 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Mechanistic Insights into the Ene-Reductase-Catalyzed Promiscuous Reduction of Oximes to Amines.
Acs Catalysis, 13, 2023
|
|
8AU8
 
 | |
8AUH
 
 | |
8AUI
 
 | |
