3HHN
 
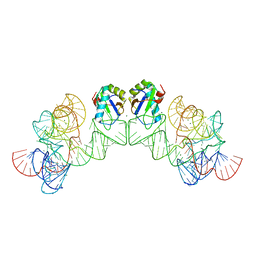 | | Crystal structure of class I ligase ribozyme self-ligation product, in complex with U1A RBD | | 分子名称: | Class I ligase ribozyme, self-ligation product, MAGNESIUM ION, ... | | 著者 | Shechner, D.M, Grant, R.A, Bagby, S.C, Bartel, D.P. | | 登録日 | 2009-05-15 | | 公開日 | 2009-11-24 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (2.987 Å) | | 主引用文献 | Crystal structure of the catalytic core of an RNA-polymerase ribozyme.
Science, 326, 2009
|
|
6VWO
 
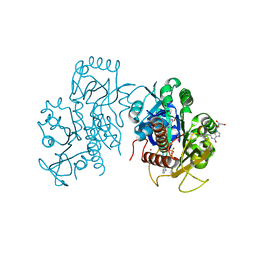 | | Crystal structure of E. coli guanosine kinase | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, GUANOSINE, Inosine-guanosine kinase, ... | | 著者 | Wang, B, Grant, R.A, Laub, M.T. | | 登録日 | 2020-02-20 | | 公開日 | 2020-10-07 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (1.78 Å) | | 主引用文献 | ppGpp Coordinates Nucleotide and Amino-Acid Synthesis in E. coli During Starvation.
Mol.Cell, 80, 2020
|
|
6VWP
 
 | | Crystal structure of E. coli guanosine kinase in complex with ppGpp | | 分子名称: | GUANOSINE, GUANOSINE-5',3'-TETRAPHOSPHATE, Inosine-guanosine kinase, ... | | 著者 | Wang, B, Grant, R.A, Laub, M.T. | | 登録日 | 2020-02-20 | | 公開日 | 2020-10-07 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (3.45 Å) | | 主引用文献 | ppGpp Coordinates Nucleotide and Amino-Acid Synthesis in E. coli During Starvation.
Mol.Cell, 80, 2020
|
|
6X0A
 
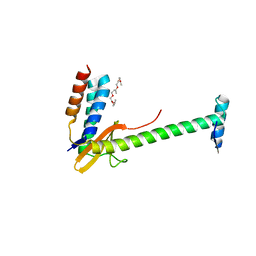 | | X-ray structure of a chimeric ParDE toxin-antitoxin complex from Mesorhizobium opportunistum | | 分子名称: | 1-ETHOXY-2-(2-ETHOXYETHOXY)ETHANE, Plasmid stabilization system, Putative addiction module antidote protein, ... | | 著者 | Lite, T.L, Grant, R.A, Laub, M.T. | | 登録日 | 2020-05-15 | | 公開日 | 2020-11-25 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | Uncovering the basis of protein-protein interaction specificity with a combinatorially complete library.
Elife, 9, 2020
|
|
3GQ0
 
 | |
3KJ0
 
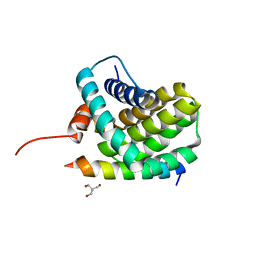 | | Mcl-1 in complex with Bim BH3 mutant I2dY | | 分子名称: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Bcl-2-like protein 11, Induced myeloid leukemia cell differentiation protein Mcl-1 | | 著者 | Fire, E, Grant, R.A, Keating, A.E. | | 登録日 | 2009-11-02 | | 公開日 | 2010-02-16 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Mcl-1-Bim complexes accommodate surprising point mutations via minor structural changes.
Protein Sci., 19, 2010
|
|
3KJ1
 
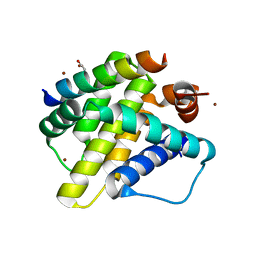 | | Mcl-1 in complex with Bim BH3 mutant I2dA | | 分子名称: | ACETATE ION, Bcl-2-like protein 11, CHLORIDE ION, ... | | 著者 | Fire, E, Grant, R.A, Keating, A.E. | | 登録日 | 2009-11-02 | | 公開日 | 2010-02-16 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (1.945 Å) | | 主引用文献 | Mcl-1-Bim complexes accommodate surprising point mutations via minor structural changes.
Protein Sci., 19, 2010
|
|
3KZ0
 
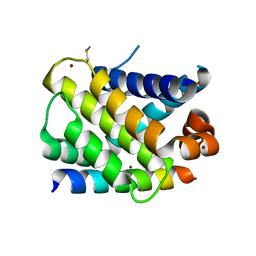 | | MCL-1 complex with MCL-1-specific selected peptide | | 分子名称: | Induced myeloid leukemia cell differentiation protein Mcl-1, Mcl-1 specific peptide MB7, SULFATE ION, ... | | 著者 | Dutta, S, Fire, E, Grant, R.A, Sauer, R.T, Keating, A.E. | | 登録日 | 2009-12-07 | | 公開日 | 2010-05-05 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (2.349 Å) | | 主引用文献 | Determinants of BH3 binding specificity for Mcl-1 versus Bcl-xL.
J.Mol.Biol., 398, 2010
|
|
3KJ2
 
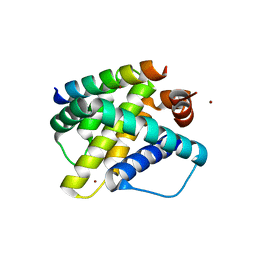 | | Mcl-1 in complex with Bim BH3 mutant F4aE | | 分子名称: | ACETATE ION, Bcl-2-like protein 11, Induced myeloid leukemia cell differentiation protein Mcl-1, ... | | 著者 | Fire, E, Grant, R.A, Keating, A.E. | | 登録日 | 2009-11-02 | | 公開日 | 2010-02-16 | | 最終更新日 | 2021-10-13 | | 実験手法 | X-RAY DIFFRACTION (2.351 Å) | | 主引用文献 | Mcl-1-Bim complexes accommodate surprising point mutations via minor structural changes.
Protein Sci., 19, 2010
|
|
3EQ2
 
 | |
3ES2
 
 | |
3F7A
 
 | |
6UA3
 
 | |
3F79
 
 | |
6UAB
 
 | |
3DNJ
 
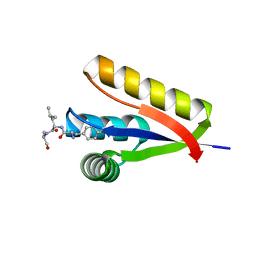 | | The structure of the Caulobacter crescentus ClpS protease adaptor protein in complex with a N-end rule peptide | | 分子名称: | ATP-dependent Clp protease adapter protein clpS, MAGNESIUM ION, synthetic N-end rule peptide | | 著者 | Wang, K, Roman-Hernandez, G, Grant, R.A, Sauer, R.T, Baker, T.A. | | 登録日 | 2008-07-02 | | 公開日 | 2008-11-18 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (1.15 Å) | | 主引用文献 | The molecular basis of N-end rule recognition.
Mol.Cell, 32, 2008
|
|
3GCO
 
 | |
3GDS
 
 | |
3GDU
 
 | |
3GCN
 
 | |
3G1B
 
 | | The structure of the M53A mutant of Caulobacter crescentus clpS protease adaptor protein in complex with WLFVQRDSKE peptide | | 分子名称: | 10-residue peptide, ATP-dependent Clp protease adapter protein clpS, MAGNESIUM ION | | 著者 | Baker, T.A, Roman-Hernandez, G, Sauer, R.T, Grant, R.A. | | 登録日 | 2009-01-29 | | 公開日 | 2009-04-28 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (1.448 Å) | | 主引用文献 | Molecular basis of substrate selection by the N-end rule adaptor protein ClpS.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
3GDV
 
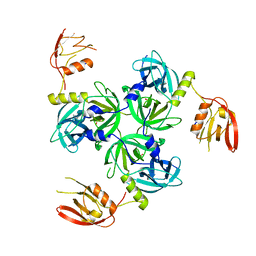 | |
3EOD
 
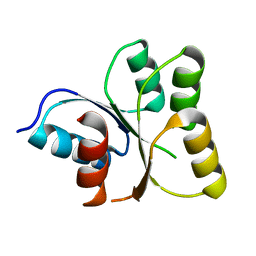 | |
3G19
 
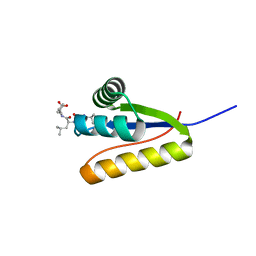 | | The structure of the Caulobacter crescentus clpS protease adaptor protein in complex with LLL tripeptide | | 分子名称: | ATP-dependent Clp protease adapter protein clpS, LLL tripeptide | | 著者 | Baker, T.A, Roman-Hernandez, G, Sauer, R.T, Grant, R.A. | | 登録日 | 2009-01-29 | | 公開日 | 2009-04-28 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (1.849 Å) | | 主引用文献 | Molecular basis of substrate selection by the N-end rule adaptor protein ClpS.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
3G3P
 
 | |
