6SAQ
 
 | | wild-type NuoEF from Aquifex aeolicus bound to NADH-OH | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, FLAVIN MONONUCLEOTIDE, GLYCEROL, ... | | Authors: | Gerhardt, S. | | Deposit date: | 2019-07-17 | | Release date: | 2020-07-22 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Structural Basis for Inhibition of ROS-Producing Respiratory Complex I by NADH-OH.
Angew.Chem.Int.Ed.Engl., 2021
|
|
2YIG
 
 | |
2JIH
 
 | | Crystal Structure of Human ADAMTS-1 catalytic Domain and Cysteine- Rich Domain (complex-form) | | Descriptor: | (2S,3R)-N~4~-[(1S)-2,2-dimethyl-1-(methylcarbamoyl)propyl]-N~1~,2-dihydroxy-3-(2-methylpropyl)butanediamide, ADAMTS-1, CADMIUM ION, ... | | Authors: | Gerhardt, S, Hassall, G, Hawtin, P, McCall, E, Flavell, L, Minshull, C, Hargreaves, D, Ting, A, Pauptit, R.A, Parker, A.E, Abbott, W.M. | | Deposit date: | 2007-06-28 | | Release date: | 2008-01-15 | | Last modified: | 2019-04-24 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structures of human ADAMTS-1 reveal a conserved catalytic domain and a disintegrin-like domain with a fold homologous to cysteine-rich domains.
J. Mol. Biol., 373, 2007
|
|
8RVC
 
 | | Crystal structure of alpha keto acid C-methyl-transferases MrsA bound to ketoarginine | | Descriptor: | 1,2-ETHANEDIOL, 2-ketoarginine methyltransferase, 5-[(diaminomethylidene)amino]-2-oxopentanoic acid, ... | | Authors: | Gerhardt, S, Kemper, F, Andexer, J.N. | | Deposit date: | 2024-02-01 | | Release date: | 2024-07-03 | | Method: | X-RAY DIFFRACTION (1.969 Å) | | Cite: | Structures and protein engineering of the alpha-keto acid C-methyltransferases SgvM and MrsA for rational substrate transfer.
Chembiochem, 2024
|
|
8RXF
 
 | |
8RVS
 
 | |
8RWW
 
 | |
8R4Z
 
 | |
8RWM
 
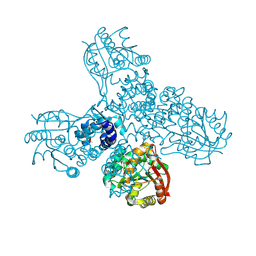 | |
2VXS
 
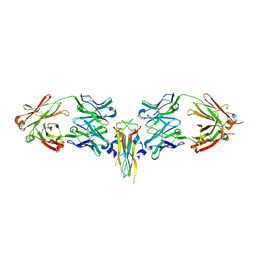 | | Structure of IL-17A in complex with a potent, fully human neutralising antibody | | Descriptor: | FAB FRAGMENT, INTERLEUKIN-17A, SULFATE ION | | Authors: | Gerhardt, S, Hargreaves, D, Pauptit, R.A, Davies, R.A, Russell, C, Welsh, F, Tuske, S.J, Coales, S.J, Hamuro, Y, Needham, M.R.C, Langham, C, Barker, W, Bell, P, Aziz, A, Smith, M.J, Dawson, S, Abbott, W.M. | | Deposit date: | 2008-07-09 | | Release date: | 2009-07-14 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.63 Å) | | Cite: | Structure of Il-17A in Complex with a Potent, Fully Human Neutralising Antibody.
J.Mol.Biol., 394, 2009
|
|
3Q2H
 
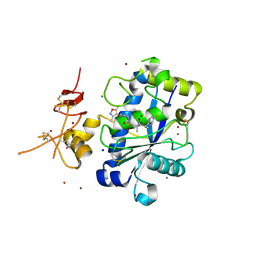 | | Adamts1 in complex with N-hydroxyformamide inhibitors of ADAM-TS4 | | Descriptor: | A disintegrin and metalloproteinase with thrombospondin motifs 1, CADMIUM ION, MAGNESIUM ION, ... | | Authors: | Gerhardt, S, Hargreaves, D. | | Deposit date: | 2010-12-20 | | Release date: | 2011-03-30 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | The design and synthesis of novel N-hydroxyformamide inhibitors of ADAM-TS4 for the treatment of osteoarthritis
Bioorg.Med.Chem.Lett., 21, 2011
|
|
3Q2G
 
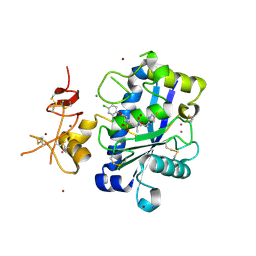 | | Adamts1 in complex with a novel N-hydroxyformamide inhibitors | | Descriptor: | A disintegrin and metalloproteinase with thrombospondin motifs 1, CADMIUM ION, MAGNESIUM ION, ... | | Authors: | Gerhardt, S, Hargreaves, D. | | Deposit date: | 2010-12-20 | | Release date: | 2011-03-30 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The design and synthesis of novel N-hydroxyformamide inhibitors of ADAM-TS4 for the treatment of osteoarthritis
Bioorg.Med.Chem.Lett., 21, 2011
|
|
2V4B
 
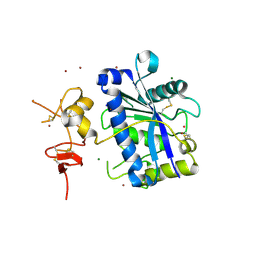 | | Crystal Structure of Human ADAMTS-1 catalytic Domain and Cysteine- Rich Domain (apo-form) | | Descriptor: | ADAMTS-1, CADMIUM ION, MAGNESIUM ION, ... | | Authors: | Gerhardt, S, Hassall, G, Hawtin, P, McCall, E, Flavell, L, Minshull, C, Hargreaves, D, Ting, A, Pauptit, R.A, Parker, A.E, Abbott, W.M. | | Deposit date: | 2007-06-28 | | Release date: | 2008-01-15 | | Last modified: | 2019-04-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structures of Human Adamts-1 Reveal a Conserved Catalytic Domain and a Disintegrin-Like Domain with a Fold Homologous to Cysteine-Rich Domains.
J.Mol.Biol., 373, 2007
|
|
6HL3
 
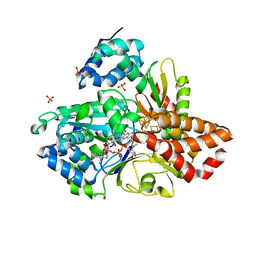 | | wild-type NuoEF from Aquifex aeolicus - oxidized form bound to NAD+ | | Descriptor: | 3[N-MORPHOLINO]PROPANE SULFONIC ACID, CHLORIDE ION, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Gerhardt, S, Friedrich, T, Einsle, O, Gnandt, E, Schulte, M, Fiegen, D. | | Deposit date: | 2018-09-10 | | Release date: | 2019-06-26 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | A mechanism to prevent production of reactive oxygen species by Escherichia coli respiratory complex I.
Nat Commun, 10, 2019
|
|
6HLM
 
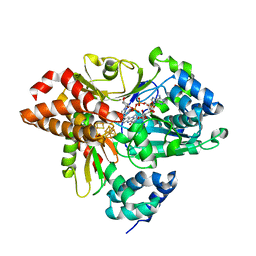 | | Variant G129D of NuoEF from Aquifex aeolicus bound to NAD+ | | Descriptor: | 3[N-MORPHOLINO]PROPANE SULFONIC ACID, FE2/S2 (INORGANIC) CLUSTER, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Gerhardt, S, Friedrich, T, Einsle, O, Gnandt, E, Schulte, M, Fiegen, D. | | Deposit date: | 2018-09-11 | | Release date: | 2019-06-26 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A mechanism to prevent production of reactive oxygen species by Escherichia coli respiratory complex I.
Nat Commun, 10, 2019
|
|
6HL2
 
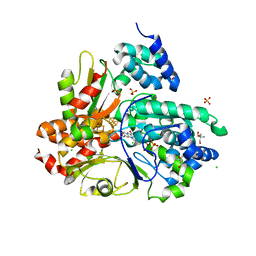 | | wild-type NuoEF from Aquifex aeolicus - oxidized form | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Gerhardt, S, Friedrich, T, Einsle, O, Gnandt, E, Schulte, M, Fiegen, D. | | Deposit date: | 2018-09-10 | | Release date: | 2019-06-26 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | A mechanism to prevent production of reactive oxygen species by Escherichia coli respiratory complex I.
Nat Commun, 10, 2019
|
|
6HLJ
 
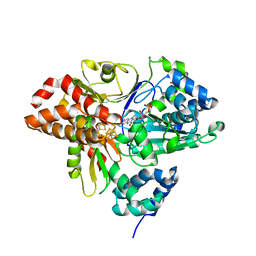 | | Variant G129S of NuoEF from Aquifex aeolicus - oxidized from | | Descriptor: | 3[N-MORPHOLINO]PROPANE SULFONIC ACID, FE2/S2 (INORGANIC) CLUSTER, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Gerhardt, S, Friedrich, T, Einsle, O, Gnandt, E, Schulte, M, Fiegen, D. | | Deposit date: | 2018-09-11 | | Release date: | 2019-06-26 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A mechanism to prevent production of reactive oxygen species by Escherichia coli respiratory complex I.
Nat Commun, 10, 2019
|
|
6HLA
 
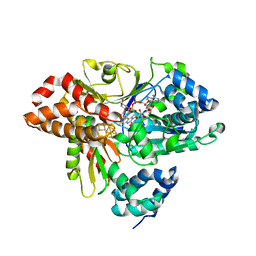 | | wild-type NuoEF from Aquifex aeolicus - reduced form bound to NADH | | Descriptor: | 3[N-MORPHOLINO]PROPANE SULFONIC ACID, FE2/S2 (INORGANIC) CLUSTER, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Gerhardt, S, Friedrich, T, Einsle, O, Gnandt, E, Schulte, M, Fiegen, D. | | Deposit date: | 2018-09-11 | | Release date: | 2019-06-26 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A mechanism to prevent production of reactive oxygen species by Escherichia coli respiratory complex I.
Nat Commun, 10, 2019
|
|
6HL4
 
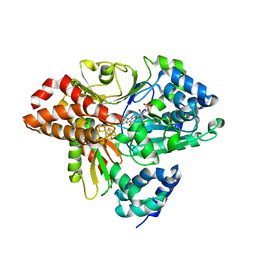 | | wild-type NuoEF from Aquifex aeolicus - reduced form | | Descriptor: | CHLORIDE ION, FE2/S2 (INORGANIC) CLUSTER, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Gerhardt, S, Friedrich, T, Einsle, O, Gnandt, E, Schulte, M, Fiegen, D. | | Deposit date: | 2018-09-10 | | Release date: | 2019-06-26 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | A mechanism to prevent production of reactive oxygen species by Escherichia coli respiratory complex I.
Nat Commun, 10, 2019
|
|
6HLI
 
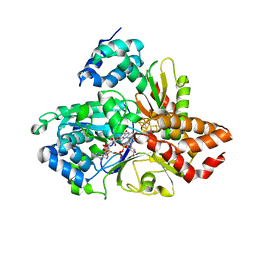 | | wild-type NuoEF from Aquifex aeolicus - reduced form bound to NAD+ | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, FLAVIN MONONUCLEOTIDE, IRON/SULFUR CLUSTER, ... | | Authors: | Gerhardt, S, Friedrich, T, Einsle, O, Gnandt, E, Schulte, M, Fiegen, D. | | Deposit date: | 2018-09-11 | | Release date: | 2019-06-26 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | A mechanism to prevent production of reactive oxygen species by Escherichia coli respiratory complex I.
Nat Commun, 10, 2019
|
|
2BAL
 
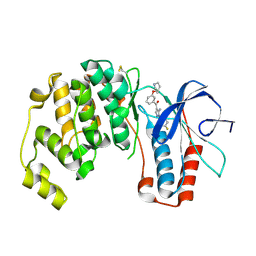 | | p38alpha MAP kinase bound to pyrazoloamine | | Descriptor: | Mitogen-activated protein kinase 14, [5-AMINO-1-(4-FLUOROPHENYL)-1H-PYRAZOL-4-YL][3-(PIPERIDIN-4-YLOXY)PHENYL]METHANONE | | Authors: | Gerhardt, S, Pauptit, R.A, Read, J, Tucker, J, Norman, R.A, Breed, J. | | Deposit date: | 2005-10-14 | | Release date: | 2005-12-06 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Prevention of MKK6-Dependent Activation by Binding to p38alpha MAP Kinase.
Biochemistry, 44, 2005
|
|
2BAQ
 
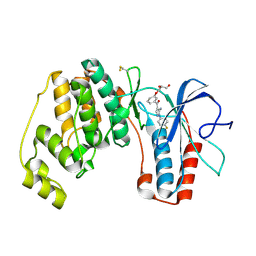 | | p38alpha bound to Ro3201195 | | Descriptor: | Mitogen-activated protein kinase 14, [5-AMINO-1-(4-FLUOROPHENYL)-1H-PYRAZOL-4-YL](3-{[(2R)-2,3-DIHYDROXYPROPYL]OXY}PHENYL)METHANONE | | Authors: | Gerhardt, S, Pauptit, R.A, Breed, J, Read, J, Tucker, J, Norman, R.A. | | Deposit date: | 2005-10-14 | | Release date: | 2005-12-06 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Prevention of MKK6-Dependent Activation by Binding to p38alpha MAP Kinase.
Biochemistry, 44, 2005
|
|
5LOG
 
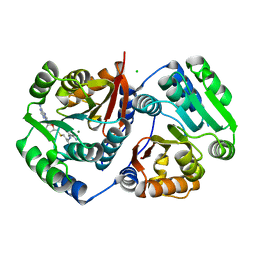 | | Crystal Structure of SafC from Myxococcus xanthus bound to SAM | | Descriptor: | CHLORIDE ION, L-DOPAMINE, MAGNESIUM ION, ... | | Authors: | Gerhardt, S, Netzer, J, Einsle, O. | | Deposit date: | 2016-08-09 | | Release date: | 2017-06-21 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Functional and structural characterisation of a bacterial O-methyltransferase and factors determining regioselectivity.
FEBS Lett., 591, 2017
|
|
4AA4
 
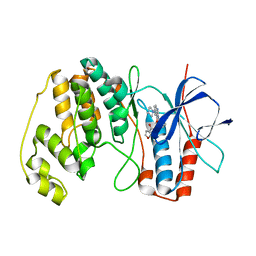 | | P38ALPHA MAP KINASE BOUND TO CMPD 22 | | Descriptor: | MITOGEN-ACTIVATED PROTEIN KINASE 14, N-[4-METHYL-3-[6-(4-METHYLPIPERAZIN-1-YL)-4-OXIDANYLIDENE-QUINAZOLIN-3-YL]PHENYL]FURAN-3-CARBOXAMIDE | | Authors: | Gerhardt, S, Hargreaves, D. | | Deposit date: | 2011-11-30 | | Release date: | 2012-05-16 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The Discovery of N-Cyclopropyl-4-Methyl-3-[6--(4-Methylpiperazin-1-Yl-4-Oxoquinazolin-3(4H)-Yl]Benzamide (Azd6703), a Clinical P38Alpha Map Kinase Inhibitor for the Treatment of Inflammatory Diseases
Bioorg.Med.Chem.Lett., 22, 2012
|
|
4A9Y
 
 | | P38ALPHA MAP KINASE BOUND TO CMPD 8 | | Descriptor: | MITOGEN-ACTIVATED PROTEIN KINASE 14, N-(3-{[7-METHOXY-6-(2-PYRROLIDIN-1-YLETHOXY)QUINAZOLIN-4-YL]AMINO}-4-METHYLPHENYL)-2-MORPHOLIN-4-YLISONICOTINAMIDE | | Authors: | Gerhardt, S, Hargreaves, D. | | Deposit date: | 2011-11-29 | | Release date: | 2012-05-16 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The Discovery of N-Cyclopropyl-4-Methyl-3-[6--(4-Methylpiperazin-1-Yl-4-Oxoquinazolin-3(4H)-Yl]Benzamide (Azd6703), a Clinical P38Alpha Map Kinase Inhibitor for the Treatment of Inflammatory Diseases
Bioorg.Med.Chem.Lett., 22, 2012
|
|
