7ROY
 
 | | The structure of the Fem1B:FNIP1 complex | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Folliculin-interacting protein 1, Protein fem-1 homolog B, ... | | Authors: | Gee, C.L, Mena, E.L, Manford, A.G, Rape, M. | | Deposit date: | 2021-08-02 | | Release date: | 2021-10-13 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural basis and regulation of the reductive stress response.
Cell, 184, 2021
|
|
2AN3
 
 | | Structure of PNMT with S-adenosyl-L-homocysteine and the semi-rigid analogue acceptor substrate cis-(1R,2S)-2-amino-1-tetralol. | | Descriptor: | CIS-(1R,2S)-2-AMINO-1,2,3,4-TETRAHYDRONAPHTHALEN-1-OL, Phenylethanolamine N-methyltransferase, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Gee, C.L, Tyndall, J.D.A, Grunewald, G.L, Wu, Q, McLeish, M.J, Martin, J.L. | | Deposit date: | 2005-08-11 | | Release date: | 2006-03-14 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Mode of binding of methyl acceptor substrates to the adrenaline-synthesizing enzyme phenylethanolamine N-methyltransferase: implications for catalysis
Biochemistry, 44, 2005
|
|
2AN5
 
 | | Structure of human PNMT complexed with S-adenosyl-homocysteine and an inhibitor, trans-(1S,2S)-2-amino-1-tetralol | | Descriptor: | PHOSPHATE ION, Phenylethanolamine N-methyltransferase, S-ADENOSYL-L-HOMOCYSTEINE, ... | | Authors: | Gee, C.L, Tyndall, J.D.A, Grunewald, G.L, Wu, Q, McLeish, M.J, Martin, J.L. | | Deposit date: | 2005-08-11 | | Release date: | 2006-03-14 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Mode of binding of methyl acceptor substrates to the adrenaline-synthesizing enzyme phenylethanolamine N-methyltransferase: implications for catalysis
Biochemistry, 44, 2005
|
|
2AN4
 
 | | Structure of PNMT complexed with S-adenosyl-L-homocysteine and the acceptor substrate octopamine | | Descriptor: | 4-(2R-AMINO-1-HYDROXYETHYL)PHENOL, PHOSPHATE ION, Phenylethanolamine N-methyltransferase, ... | | Authors: | Gee, C.L, Tyndall, J.D.A, Grunewald, G.L, Wu, Q, McLeish, M.J, Martin, J.L. | | Deposit date: | 2005-08-11 | | Release date: | 2006-03-14 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Mode of binding of methyl acceptor substrates to the adrenaline-synthesizing enzyme phenylethanolamine N-methyltransferase: implications for catalysis
Biochemistry, 44, 2005
|
|
8UH7
 
 | | Structure of T4 Bacteriophage clamp loader bound to the T4 clamp, primer-template DNA, and ATP analog | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Primer DNA strand, ... | | Authors: | Gee, C.L, Marcus, K, Kelch, B.A, Makino, D.L. | | Deposit date: | 2023-10-07 | | Release date: | 2023-12-13 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.628 Å) | | Cite: | Autoinhibition of a clamp-loader ATPase revealed by deep mutagenesis and cryo-EM.
Nat.Struct.Mol.Biol., 31, 2024
|
|
7SOJ
 
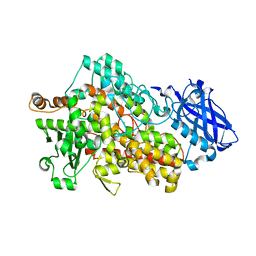 | |
7SOI
 
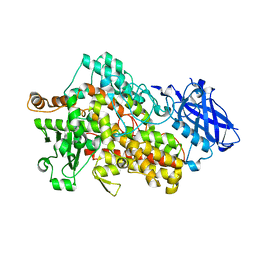 | | Structure of I552A Soybean Lipoxygenase at 277K | | Descriptor: | FE (III) ION, Lipoxygenase, SODIUM ION | | Authors: | Gee, C.L, Offenbacher, A.R, Hu, S. | | Deposit date: | 2021-10-31 | | Release date: | 2022-11-09 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Temporal and spatial resolution of distal protein motions that activate hydrogen tunneling in soybean lipoxygenase.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
3OUK
 
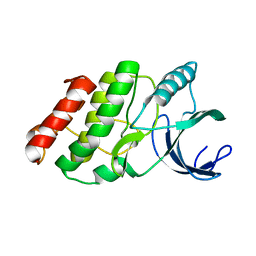 | |
3OTV
 
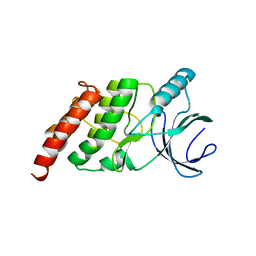 | |
3OUN
 
 | |
3SBO
 
 | | Structure of E.coli GDH from native source | | Descriptor: | CHLORIDE ION, NADP-specific glutamate dehydrogenase | | Authors: | Gee, C.L, Zubieta, C, Echols, N, Totir, M. | | Deposit date: | 2011-06-06 | | Release date: | 2012-03-21 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.204 Å) | | Cite: | Macro-to-Micro Structural Proteomics: Native Source Proteins for High-Throughput Crystallization.
Plos One, 7, 2012
|
|
3UQC
 
 | |
6OF9
 
 | |
6OF8
 
 | | Structure of Thr354Asn, Glu355Gln, Thr412Asn, Ile414Met, Ile464His, and Phe467Met mutant human CamKII-alpha hub domain | | Descriptor: | Calcium/calmodulin-dependent protein kinase type II subunit alpha, GLYCEROL, POTASSIUM ION | | Authors: | McSpadden, E.D, Chi, C.C, Gee, C.L, Kuriyan, J. | | Deposit date: | 2019-03-28 | | Release date: | 2019-04-17 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Variation in assembly stoichiometry in non-metazoan homologs of the hub domain of Ca2+/calmodulin-dependent protein kinase II.
Protein Sci., 28, 2019
|
|
7REC
 
 | | Structure of Thr354Asn, Glu355Gln, Thr412Asn, Ile414Met, Ile464His, and Phe467Met mutant human CaMKII alpha hub bound to 5-HDC | | Descriptor: | 5-hydroxydiclofenac, Calcium/calmodulin-dependent protein kinase type II subunit alpha, SODIUM ION | | Authors: | McSpadden, E.D, Chi, C.C, Gee, C.L, Kuriyan, J. | | Deposit date: | 2021-07-12 | | Release date: | 2021-07-21 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | GHB analogs confer neuroprotection through specific interaction with the CaMKII alpha hub domain.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
1YZ3
 
 | | Structure of human pnmt complexed with cofactor product adohcy and inhibitor SK&F 64139 | | Descriptor: | 7,8-DICHLORO-1,2,3,4-TETRAHYDROISOQUINOLINE, Phenylethanolamine N-methyltransferase, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Wu, Q, Gee, C.L, Lin, F, Martin, J.L, Grunewald, G.L, McLeish, M.J. | | Deposit date: | 2005-02-27 | | Release date: | 2006-02-07 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural, mutagenic, and kinetic analysis of the binding of substrates and inhibitors of human phenylethanolamine N-methyltransferase
J.Med.Chem., 48, 2005
|
|
7U0R
 
 | | Crystal structure of Methanomethylophilus alvus PylRS(N166A/V168A) complexed with meta-trifluoromethyl-2-benzylmalonate and AMP-PNP | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Swenson, C.V, Roe, L.T, Fricke, R.C.B, Smaga, S.S, Gee, C.L, Schepartz, A. | | Deposit date: | 2022-02-18 | | Release date: | 2023-06-14 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Expanding the substrate scope of pyrrolysyl-transfer RNA synthetase enzymes to include non-alpha-amino acids in vitro and in vivo.
Nat.Chem., 15, 2023
|
|
5WDQ
 
 | | H-Ras mutant L120A bound to GMP-PNP at 100K | | Descriptor: | ACETATE ION, CALCIUM ION, GTPase HRas, ... | | Authors: | Bandaru, P, Gee, C.L, Kuriyan, J. | | Deposit date: | 2017-07-05 | | Release date: | 2017-07-19 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Deconstruction of the Ras switching cycle through saturation mutagenesis.
Elife, 6, 2017
|
|
5WDR
 
 | | Choanoflagellate Salpingoeca rosetta Ras with GMP-PNP | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, Ras protein, ... | | Authors: | Kondo, Y, Gee, C.L, Kuriyan, J. | | Deposit date: | 2017-07-05 | | Release date: | 2017-07-19 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Deconstruction of the Ras switching cycle through saturation mutagenesis.
Elife, 6, 2017
|
|
5WDP
 
 | | H-Ras mutant L120A bound to GMP-PNP at 277K | | Descriptor: | CALCIUM ION, GTPase HRas, MAGNESIUM ION, ... | | Authors: | Cofsky, J.C, Bandaru, P, Gee, C.L, Kuriyan, J. | | Deposit date: | 2017-07-05 | | Release date: | 2017-07-19 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Deconstruction of the Ras switching cycle through saturation mutagenesis.
Elife, 6, 2017
|
|
9EOY
 
 | | Structure of Thr354Asn, Glu355Gln, Thr412Asn, Ile414Met, Ile464His, and Phe467Met mutant human CaMKII alpha hub bound to PIPA | | Descriptor: | 2-[6-(4-chlorophenyl)imidazo[1,2-b]pyridazin-2-yl]ethanoic acid, ACETATE ION, Calcium/calmodulin-dependent protein kinase type II subunit alpha, ... | | Authors: | Narayanan, D, Larsen, A.S.G, Solbak, S.M.O, Wellendorph, P, Gee, C.L, Kastrup, J.S. | | Deposit date: | 2024-03-15 | | Release date: | 2024-09-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Ligand-induced CaMKII alpha hub Trp403 flip, hub domain stacking, and modulation of kinase activity.
Protein Sci., 33, 2024
|
|
2XHY
 
 | | Crystal Structure of E.coli BglA | | Descriptor: | 6-PHOSPHO-BETA-GLUCOSIDASE BGLA, BROMIDE ION, SULFATE ION | | Authors: | Totir, M, Zubieta, C, Echols, N, May, A.P, Gee, C.L, nanao, M, alber, T. | | Deposit date: | 2010-06-24 | | Release date: | 2011-07-06 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Macro-to-Micro Structural Proteomics: Native Source Proteins for High-Throughput Crystallization.
Plos One, 7, 2012
|
|
2OZ5
 
 | | Crystal structure of Mycobacterium tuberculosis protein tyrosine phosphatase PtpB in complex with the specific inhibitor OMTS | | Descriptor: | Phosphotyrosine protein phosphatase ptpb, {(3-CHLOROBENZYL)[(5-{[(3,3-DIPHENYLPROPYL)AMINO]SULFONYL}-2-THIENYL)METHYL]AMINO}(OXO)ACETIC ACID | | Authors: | Grundner, C, Gee, C.L, Alber, T, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2007-02-23 | | Release date: | 2007-05-01 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Basis for Selective Inhibition of Mycobacterium tuberculosis Protein Tyrosine Phosphatase PtpB.
Structure, 15, 2007
|
|
8UK9
 
 | | Structure of T4 Bacteriophage clamp loader mutant D110C bound to the T4 clamp, primer-template DNA, and ATP analog | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ALUMINUM FLUORIDE, DNA primer, ... | | Authors: | Marcus, K, Ghaffari-Kashani, S, Gee, C.L. | | Deposit date: | 2023-10-12 | | Release date: | 2023-12-13 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Autoinhibition of a clamp-loader ATPase revealed by deep mutagenesis and cryo-EM.
Nat.Struct.Mol.Biol., 31, 2024
|
|
7SA7
 
 | |
