1PWE
 
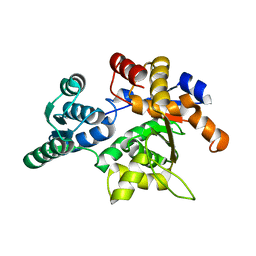 | | Rat Liver L-Serine Dehydratase Apo Enzyme | | Descriptor: | L-serine dehydratase | | Authors: | Yamada, T, Komoto, J, Takata, Y, Ogawa, H, Takusagawa, F. | | Deposit date: | 2003-07-01 | | Release date: | 2003-12-02 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of serine dehydratase from rat liver.
Biochemistry, 42, 2003
|
|
1XWF
 
 | | K185N mutated S-adenosylhomocysteine hydrolase | | Descriptor: | ADENOSINE, Adenosylhomocysteinase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Yamada, T, Takata, Y, Komoto, J, Gomi, T, Ogawa, H, Fujioka, M, Takusagawa, F. | | Deposit date: | 2004-11-01 | | Release date: | 2005-09-20 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Catalytic mechanism of S-adenosylhomocysteine hydrolase: Roles of His 54, Asp130, Glu155, Lys185, and Aspl89.
Int.J.Biochem.Cell Biol., 37, 2005
|
|
1XVA
 
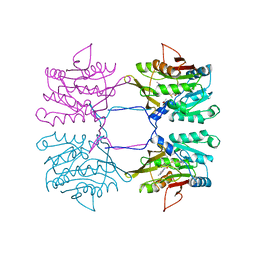 | | METHYLTRANSFERASE | | Descriptor: | ACETATE ION, GLYCINE N-METHYLTRANSFERASE, S-ADENOSYLMETHIONINE | | Authors: | Fu, Z, Hu, Y, Konishi, K, Takata, Y, Ogawa, H, Gomi, T, Fujioka, M, Takusagawa, F. | | Deposit date: | 1996-07-20 | | Release date: | 1997-01-27 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of glycine N-methyltransferase from rat liver.
Biochemistry, 35, 1996
|
|
1C7Y
 
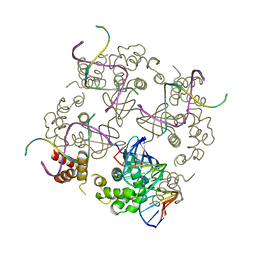 | | E.COLI RUVA-HOLLIDAY JUNCTION COMPLEX | | Descriptor: | DNA (5'-D(P*DAP*DAP*DGP*DTP*DTP*DGP*DGP*DGP*DAP*DTP*DTP*DGP*DT)-3'), DNA (5'-D(P*DCP*DAP*DAP*DTP*DCP*DCP*DCP*DAP*DAP*DCP*DTP*DT)-3'), DNA (5'-D(P*DCP*DGP*DAP*DAP*DTP*DGP*DTP*DGP*DTP*DGP*DTP*DCP*DT)-3'), ... | | Authors: | Ariyoshi, M, Nishino, T, Iwasaki, H, Shinagawa, H, Morikawa, K. | | Deposit date: | 2000-04-03 | | Release date: | 2000-07-21 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Crystal structure of the holliday junction DNA in complex with a single RuvA tetramer.
Proc.Natl.Acad.Sci.USA, 97, 2000
|
|
2RKB
 
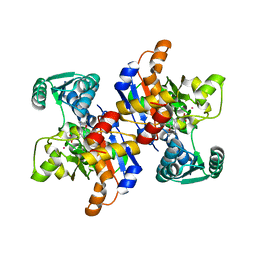 | | Serine dehydratase like-1 from human cancer cells | | Descriptor: | POTASSIUM ION, PYRIDOXAL-5'-PHOSPHATE, Serine dehydratase-like | | Authors: | Yamada, T, Komoto, J, Kasuya, T, Mori, H, Ogawa, H, Takusagawa, F. | | Deposit date: | 2007-10-16 | | Release date: | 2008-04-01 | | Last modified: | 2017-10-25 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | A catalytic mechanism that explains a low catalytic activity of serine dehydratase like-1 from human cancer cells: Crystal structure and site-directed mutagenesis studies.
Biochim.Biophys.Acta, 1780, 2008
|
|
1BXU
 
 | | OXIDIZED PLASTOCYANIN FROM SYNECHOCOCCUS SP. | | Descriptor: | COPPER (II) ION, PLASTOCYANIN | | Authors: | Inoue, T, Sugawara, H, Hamanaka, S, Tsukui, H, Suzuki, E, Kohzuma, T, Kai, Y. | | Deposit date: | 1998-10-09 | | Release date: | 1999-06-15 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure determinations of oxidized and reduced plastocyanin from the cyanobacterium Synechococcus sp. PCC 7942.
Biochemistry, 38, 1999
|
|
1D8L
 
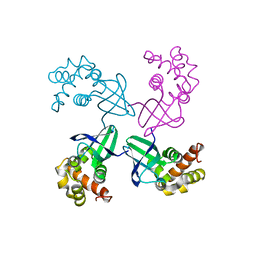 | | E. COLI HOLLIDAY JUNCTION BINDING PROTEIN RUVA NH2 REGION LACKING DOMAIN III | | Descriptor: | PROTEIN (HOLLIDAY JUNCTION DNA HELICASE RUVA) | | Authors: | Nishino, T, Iwasaki, H, Kataoka, M, Ariyoshi, M, Fujita, T, Shinagawa, H, Morikawa, K. | | Deposit date: | 1999-10-25 | | Release date: | 2000-05-03 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Modulation of RuvB function by the mobile domain III of the Holliday junction recognition protein RuvA.
J.Mol.Biol., 298, 2000
|
|
1CP8
 
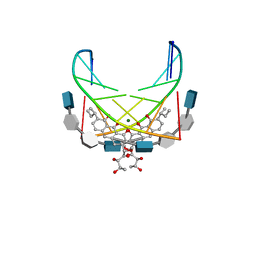 | | NMR STRUCTURE OF DNA (5'-D(TTGGCCAA)2-3') COMPLEXED WITH NOVEL ANTITUMOR DRUG UCH9 | | Descriptor: | 1,2-HYDRO-1-OXY-3,4-HYDRO-3-(1-METHOXY-2-OXY-3,4-DIHYDROXYPENTYL)-8,9-DIHYDROXY-7-(SEC-BUTYL)-ANTHRACENE, DNA (5'-D(P*TP*TP*GP*GP*CP*CP*AP*A)-3'), MAGNESIUM ION, ... | | Authors: | Katahira, R, Katahira, M, Yamashita, Y, Ogawa, H, Kyogoku, Y, Yoshida, M. | | Deposit date: | 1999-06-11 | | Release date: | 1999-07-01 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the novel antitumor drug UCH9 complexed with d(TTGGCCAA)2 as determined by NMR.
Nucleic Acids Res., 26, 1998
|
|
3VX7
 
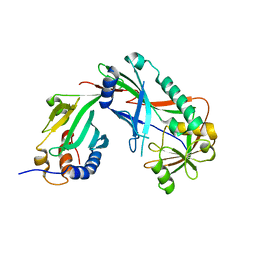 | | Crystal structure of Kluyveromyces marxianus Atg7NTD-Atg10 complex | | Descriptor: | E1, E2 | | Authors: | Yamaguchi, M, Matoba, K, Sawada, R, Fujioka, Y, Nakatogawa, H, Yamamoto, H, Kobashigawa, Y, Hoshida, H, Akada, R, Ohsumi, Y, Noda, N.N, Inagaki, F. | | Deposit date: | 2012-09-11 | | Release date: | 2012-11-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Noncanonical recognition and UBL loading of distinct E2s by autophagy-essential Atg7.
Nat.Struct.Mol.Biol., 19, 2012
|
|
3VX6
 
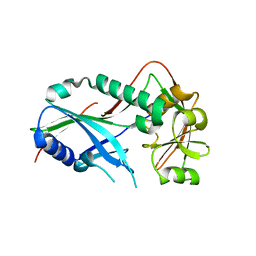 | | Crystal structure of Kluyveromyces marxianus Atg7NTD | | Descriptor: | E1 | | Authors: | Yamaguchi, M, Matoba, K, Sawada, R, Fujioka, Y, Nakatogawa, H, Yamamoto, H, Kobashigawa, Y, Hoshida, H, Akada, R, Ohsumi, Y, Noda, N.N, Inagaki, F. | | Deposit date: | 2012-09-11 | | Release date: | 2012-11-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Noncanonical recognition and UBL loading of distinct E2s by autophagy-essential Atg7.
Nat.Struct.Mol.Biol., 19, 2012
|
|
7VR5
 
 | | Crystal structure of CmABCB1 W114Y/W161Y/W363Y/W364Y/M391W (4WY/M391W) mutant | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, DECYL-BETA-D-MALTOPYRANOSIDE, Probable ATP-dependent transporter ycf16 | | Authors: | Inoue, Y, Ogawa, H, Kato, H. | | Deposit date: | 2021-10-21 | | Release date: | 2022-09-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure-based alteration of tryptophan residues of the multidrug transporter CmABCB1 to assess substrate binding using fluorescence spectroscopy.
Protein Sci., 31, 2022
|
|
7VMM
 
 | | Structure of recombinant RyR2 (EGTA dataset, class 1, closed state) | | Descriptor: | Peptidyl-prolyl cis-trans isomerase FKBP1B, Ryanodine receptor 2, ZINC ION | | Authors: | Kobayashi, T, Tsutsumi, A, Kurebayashi, N, Kodama, M, Kikkawa, M, Murayama, T, Ogawa, H. | | Deposit date: | 2021-10-09 | | Release date: | 2022-08-10 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Molecular basis for gating of cardiac ryanodine receptor explains the mechanisms for gain- and loss-of function mutations.
Nat Commun, 13, 2022
|
|
7VML
 
 | | Structure of recombinant RyR2 (EGTA dataset, class 1&2, closed state) | | Descriptor: | Peptidyl-prolyl cis-trans isomerase FKBP1B, Ryanodine receptor 2, ZINC ION | | Authors: | Kobayashi, T, Tsutsumi, A, Kurebayashi, N, Kodama, M, Kikkawa, M, Murayama, T, Ogawa, H. | | Deposit date: | 2021-10-09 | | Release date: | 2022-08-10 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Molecular basis for gating of cardiac ryanodine receptor explains the mechanisms for gain- and loss-of function mutations.
Nat Commun, 13, 2022
|
|
7VMN
 
 | | Structure of recombinant RyR2 (EGTA dataset, class 2, closed state) | | Descriptor: | Peptidyl-prolyl cis-trans isomerase FKBP1B, Ryanodine receptor 2, ZINC ION | | Authors: | Kobayashi, T, Tsutsumi, A, Kurebayashi, N, Kodama, M, Kikkawa, M, Murayama, T, Ogawa, H. | | Deposit date: | 2021-10-09 | | Release date: | 2022-08-10 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Molecular basis for gating of cardiac ryanodine receptor explains the mechanisms for gain- and loss-of function mutations.
Nat Commun, 13, 2022
|
|
7VMO
 
 | | Structure of recombinant RyR2 (Ca2+ dataset, class 1, open state) | | Descriptor: | CALCIUM ION, Peptidyl-prolyl cis-trans isomerase FKBP1B, Ryanodine receptor 2, ... | | Authors: | Kobayashi, T, Tsutsumi, A, Kurebayashi, N, Kodama, M, Kikkawa, M, Murayama, T, Ogawa, H. | | Deposit date: | 2021-10-09 | | Release date: | 2022-08-10 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Molecular basis for gating of cardiac ryanodine receptor explains the mechanisms for gain- and loss-of function mutations.
Nat Commun, 13, 2022
|
|
7VMP
 
 | | Structure of recombinant RyR2 (Ca2+ dataset, class 2, open state) | | Descriptor: | CALCIUM ION, Peptidyl-prolyl cis-trans isomerase FKBP1B, Ryanodine receptor 2, ... | | Authors: | Kobayashi, T, Tsutsumi, A, Kurebayashi, N, Kodama, M, Kikkawa, M, Murayama, T, Ogawa, H. | | Deposit date: | 2021-10-09 | | Release date: | 2022-08-10 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Molecular basis for gating of cardiac ryanodine receptor explains the mechanisms for gain- and loss-of function mutations.
Nat Commun, 13, 2022
|
|
7VMQ
 
 | | Structure of recombinant RyR2 (Ca2+ dataset, class 3, open state) | | Descriptor: | CALCIUM ION, Peptidyl-prolyl cis-trans isomerase FKBP1B, Ryanodine receptor 2, ... | | Authors: | Kobayashi, T, Tsutsumi, A, Kurebayashi, N, Kodama, M, Kikkawa, M, Murayama, T, Ogawa, H. | | Deposit date: | 2021-10-09 | | Release date: | 2022-08-10 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Molecular basis for gating of cardiac ryanodine receptor explains the mechanisms for gain- and loss-of function mutations
Nat Commun, 13, 2022
|
|
7VMS
 
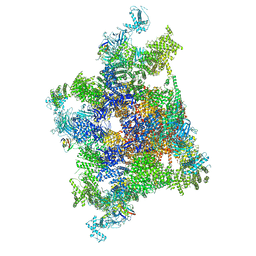 | | Structure of recombinant RyR2 mutant K4593A (Ca2+ dataset) | | Descriptor: | CALCIUM ION, Peptidyl-prolyl cis-trans isomerase FKBP1B, Ryanodine receptor 2, ... | | Authors: | Kobayashi, T, Tsutsumi, A, Kurebayashi, N, Kodama, M, Kikkawa, M, Murayama, T, Ogawa, H. | | Deposit date: | 2021-10-09 | | Release date: | 2022-08-10 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Molecular basis for gating of cardiac ryanodine receptor explains the mechanisms for gain- and loss-of function mutations
Nat Commun, 13, 2022
|
|
7VMR
 
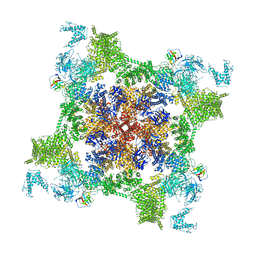 | | Structure of recombinant RyR2 mutant K4593A (EGTA dataset) | | Descriptor: | Peptidyl-prolyl cis-trans isomerase FKBP1B, Ryanodine receptor 2, ZINC ION | | Authors: | Kobayashi, T, Tsutsumi, A, Kurebayashi, N, Kodama, M, Kikkawa, M, Murayama, T, Ogawa, H. | | Deposit date: | 2021-10-09 | | Release date: | 2022-08-10 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Molecular basis for gating of cardiac ryanodine receptor explains the mechanisms for gain- and loss-of function mutations.
Nat Commun, 13, 2022
|
|
7PY2
 
 | | Structure of pathological TDP-43 filaments from ALS with FTLD | | Descriptor: | TAR DNA-binding protein 43 | | Authors: | Arseni, D, Hasegawa, H, Murzin, A.G, Kametani, F, Arai, M, Yoshida, M, Falcon, B. | | Deposit date: | 2021-10-08 | | Release date: | 2021-12-15 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (2.59 Å) | | Cite: | Structure of pathological TDP-43 filaments from ALS with FTLD.
Nature, 601, 2022
|
|
1BXV
 
 | | REDUCED PLASTOCYANIN FROM SYNECHOCOCCUS SP. | | Descriptor: | COPPER (II) ION, PLASTOCYANIN | | Authors: | Inoue, T, Sugawara, H, Hamanaka, S, Tsukui, H, Suzuki, E, Kohzuma, T, Kai, Y. | | Deposit date: | 1998-10-09 | | Release date: | 1999-06-15 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure determinations of oxidized and reduced plastocyanin from the cyanobacterium Synechococcus sp. PCC 7942.
Biochemistry, 38, 1999
|
|
5ZR0
 
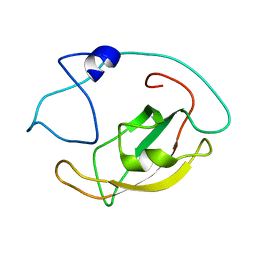 | | Solution structure of peptidyl-prolyl cis/trans isomerase domain of Trigger Factor in complex with MBP | | Descriptor: | Maltose-binding periplasmic protein,Trigger factor | | Authors: | Kawagoe, S, Nakagawa, H, Kumeta, H, Ishimori, K, Saio, T. | | Deposit date: | 2018-04-21 | | Release date: | 2018-08-22 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural insight into prolinecis/transisomerization of unfolded proteins catalyzed by the trigger factor chaperone.
J. Biol. Chem., 293, 2018
|
|
5XFA
 
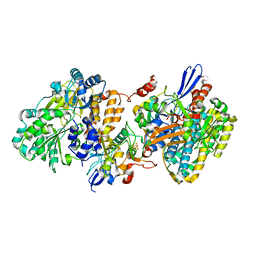 | | Crystal structure of NAD+-reducing [NiFe]-hydrogenase in the H2-reduced state | | Descriptor: | CARBONMONOXIDE-(DICYANO) IRON, FE2/S2 (INORGANIC) CLUSTER, IRON/SULFUR CLUSTER, ... | | Authors: | Shomura, Y, Taketa, M, Nakashima, H, Tai, H, Nakagawa, H, Ikeda, Y, Ishii, M, Igarashi, Y, Nishihara, H, Yoon, K.S, Ogo, S, Hirota, S, Higuchi, Y. | | Deposit date: | 2017-04-09 | | Release date: | 2017-08-23 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis of the redox switches in the NAD(+)-reducing soluble [NiFe]-hydrogenase
Science, 357, 2017
|
|
2K6Q
 
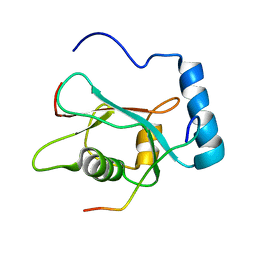 | | LC3 p62 complex structure | | Descriptor: | Microtubule-associated proteins 1A/1B light chain 3B, p62_peptide from Sequestosome-1 | | Authors: | Noda, N, Kumeta, H, Nakatogawa, H, Satoo, K, Adachi, W, Ishii, J, Fujioka, Y, Ohsumi, Y, Inagaki, F. | | Deposit date: | 2008-07-17 | | Release date: | 2008-09-02 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural basis of target recognition by ATG8/LC3 during selective autophagy
To be Published
|
|
2KWC
 
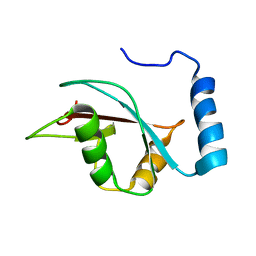 | | The NMR structure of the autophagy-related protein Atg8 | | Descriptor: | Autophagy-related protein 8 | | Authors: | Kumeta, H, Watanabe, M, Nakatogawa, H, Yamaguchi, M, Ogura, K, Adachi, W, Fujioka, Y, Noda, N.N, Ohsumi, Y, Inagaki, F. | | Deposit date: | 2010-04-05 | | Release date: | 2010-05-12 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The NMR structure of the autophagy-related protein Atg8
J.Biomol.Nmr, 47, 2010
|
|
