5ZWY
 
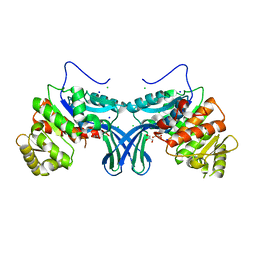 | | Ribokinase from Leishmania donovani | | Descriptor: | CHLORIDE ION, GLYCEROL, Ribokinase, ... | | Authors: | Gatreddi, S, Vasudevan, D, Qureshi, I.A. | | Deposit date: | 2018-05-17 | | Release date: | 2019-05-22 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Unraveling structural insights of ribokinase from Leishmania donovani.
Int.J.Biol.Macromol., 136, 2019
|
|
6A8C
 
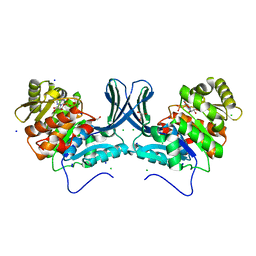 | | Ribokinase from Leishmania donovani with ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CHLORIDE ION, GLYCEROL, ... | | Authors: | Gatreddi, S, Vasudevan, D, Qureshi, I.A. | | Deposit date: | 2018-07-06 | | Release date: | 2019-07-10 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Unraveling structural insights of ribokinase from Leishmania donovani.
Int.J.Biol.Macromol., 136, 2019
|
|
6A8A
 
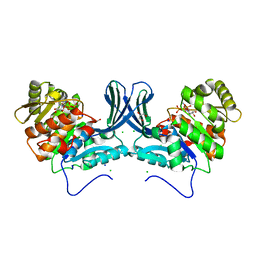 | | Ribokinase from Leishmania donovani with ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CHLORIDE ION, GLYCEROL, ... | | Authors: | Gatreddi, S, Qureshi, I.A. | | Deposit date: | 2018-07-06 | | Release date: | 2019-07-10 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Unraveling structural insights of ribokinase from Leishmania donovani.
Int.J.Biol.Macromol., 136, 2019
|
|
1I7E
 
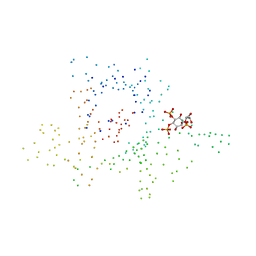 | | C-Terminal Domain Of Mouse Brain Tubby Protein bound to Phosphatidylinositol 4,5-bis-phosphate | | Descriptor: | L-ALPHA-GLYCEROPHOSPHO-D-MYO-INOSITOL-4,5-BIS-PHOSPHATE, TUBBY PROTEIN | | Authors: | Santagata, S, Boggon, T.J, Baird, C.L, Shan, W.S, Shapiro, L. | | Deposit date: | 2001-03-08 | | Release date: | 2001-06-27 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | G-protein signaling through tubby proteins.
Science, 292, 2001
|
|
7S91
 
 | |
2BD0
 
 | | Chlorobium tepidum Sepiapterin Reductase complexed with NADP and Sepiapterin | | Descriptor: | BIOPTERIN, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, sepiapterin reductase | | Authors: | Supangat, S, Seo, K.H, Choi, Y.K, Park, Y.S, Lee, K.H. | | Deposit date: | 2005-10-19 | | Release date: | 2005-11-29 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure of Chlorobium tepidum sepiapterin reductase complex reveals the novel substrate binding mode for stereospecific production of L-threo-tetrahydrobiopterin
J.Biol.Chem., 281, 2006
|
|
8EZF
 
 | | A tethered niacin-derived pincer complex with a nickel-carbon or sulfite-carbon bond in lactate racemase R98A/R100A variant | | Descriptor: | (4S)-5-methanethioyl-1-(5-O-phosphono-beta-D-ribofuranosyl)-4-sulfo-1,4-dihydropyridine-3-carbothioic S-acid, 1,2-ETHANEDIOL, CALCIUM ION, ... | | Authors: | Gatreddi, S, Hausinger, R.P, Hu, J. | | Deposit date: | 2022-10-31 | | Release date: | 2023-01-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Irreversible inactivation of lactate racemase by sodium borohydride reveals reactivity of the nickel-pincer nucleotide cofactor.
Acs Catalysis, 13, 2023
|
|
8EZH
 
 | | A tethered niacin-derived pincer complex with a nickel-carbon bond in lactate racemase R98A/R100A variant modeled with sulfite-NPN adduct | | Descriptor: | (4S)-5-methanethioyl-1-(5-O-phosphono-beta-D-ribofuranosyl)-4-sulfo-1,4-dihydropyridine-3-carbothioic S-acid, 1,2-ETHANEDIOL, CALCIUM ION, ... | | Authors: | Gatreddi, S, Hausinger, R.P, Hu, J. | | Deposit date: | 2022-10-31 | | Release date: | 2023-01-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Irreversible inactivation of lactate racemase by sodium borohydride reveals reactivity of the nickel-pincer nucleotide cofactor.
Acs Catalysis, 13, 2023
|
|
8EZI
 
 | | A tethered niacin-derived pincer complex with a nickel-carbon bond in lactate racemase R98A/R100A variant modeled with separated sulfite and NPN | | Descriptor: | 1,2-ETHANEDIOL, 3-methanethioyl-1-(5-O-phosphono-beta-D-ribofuranosyl)-5-(sulfanylcarbonyl)pyridin-1-ium, CALCIUM ION, ... | | Authors: | Gatreddi, S, Hausinger, R.P, Hu, J. | | Deposit date: | 2022-10-31 | | Release date: | 2023-01-25 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Irreversible inactivation of lactate racemase by sodium borohydride reveals reactivity of the nickel-pincer nucleotide cofactor.
Acs Catalysis, 13, 2023
|
|
5VJH
 
 | | Closed State CryoEM Reconstruction of Hsp104:ATPyS and FITC casein | | Descriptor: | FITC casein, Heat shock protein 104, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER | | Authors: | Gates, S.N, Yokom, A.L, Lin, J.-B, Jackrel, M.E, Rizo, A.N, Kendsersky, N.M, Buell, C.E, Sweeny, E.A, Chuang, E, Torrente, M.P, Mack, K.L, Su, M, Shorter, J, Southworth, D.R. | | Deposit date: | 2017-04-19 | | Release date: | 2017-07-05 | | Last modified: | 2017-08-02 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Ratchet-like polypeptide translocation mechanism of the AAA+ disaggregase Hsp104.
Science, 357, 2017
|
|
5VYA
 
 | | S. cerevisiae Hsp104:casein complex, Extended Conformation | | Descriptor: | Alpha-S1-casein, Heat shock protein 104, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER | | Authors: | Gates, S.N, Yokom, A.L, Lin, J.-B, Jackrel, M.E, Rizo, A.N, Kendsersky, N.M, Buell, C.E, Sweeny, E.A, Chuang, E, Torrente, M.P, Mack, K.L, Su, M, Shorter, J, Southworth, D.R. | | Deposit date: | 2017-05-24 | | Release date: | 2017-07-05 | | Last modified: | 2017-08-02 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Ratchet-like polypeptide translocation mechanism of the AAA+ disaggregase Hsp104.
Science, 357, 2017
|
|
5VY9
 
 | | S. cerevisiae Hsp104:casein complex, Middle Domain Conformation | | Descriptor: | Alpha-S1-casein, Heat shock protein 104, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER | | Authors: | Gates, S.N, Yokom, A.L, Lin, J.-B, Jackrel, M.E, Rizo, A.N, Kendsersky, N.M, Buell, C.E, Sweeny, E.A, Chuang, E, Torrente, M.P, Mack, K.L, Su, M, Shorter, J, Southworth, D.R. | | Deposit date: | 2017-05-24 | | Release date: | 2017-07-19 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (6.7 Å) | | Cite: | Ratchet-like polypeptide translocation mechanism of the AAA+ disaggregase Hsp104.
Science, 357, 2017
|
|
5VY8
 
 | | S. cerevisiae Hsp104-ADP complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Heat shock protein 104 | | Authors: | Gates, S.N, Yokom, A.L, Lin, J.-B, Jackrel, M.E, Rizo, A.N, Kendsersky, N.M, Buell, C.E, Sweeny, E.A, Chuang, E, Torrente, M.P, Mack, K.L, Su, M, Shorter, J, Southworth, D.R. | | Deposit date: | 2017-05-24 | | Release date: | 2017-07-05 | | Last modified: | 2018-08-15 | | Method: | ELECTRON MICROSCOPY (5.6 Å) | | Cite: | Ratchet-like polypeptide translocation mechanism of the AAA+ disaggregase Hsp104.
Science, 357, 2017
|
|
6A8B
 
 | | Ribokinase from Leishmania donovani with AMPPCP | | Descriptor: | CHLORIDE ION, GLYCEROL, PHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER, ... | | Authors: | Gatreddi, S, Pillalamarri, V, Qureshi, I.A. | | Deposit date: | 2018-07-06 | | Release date: | 2019-07-10 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Unraveling structural insights of ribokinase from Leishmania donovani.
Int.J.Biol.Macromol., 136, 2019
|
|
1NKW
 
 | | Crystal Structure Of The Large Ribosomal Subunit From Deinococcus Radiodurans | | Descriptor: | 23S ribosomal RNA, 50S ribosomal protein L11, 50S ribosomal protein L13, ... | | Authors: | Harms, J.M, Schluenzen, F, Zarivach, R, Bashan, A, Gat, S, Agmon, I, Bartels, H, Franceschi, F, Yonath, A. | | Deposit date: | 2003-01-05 | | Release date: | 2003-02-11 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | High resolution structure of the large ribosomal subunit from a mesophilic eubacterium
Cell(Cambridge,Mass.), 107, 2001
|
|
6OAY
 
 | | Structure of the hyperactive ClpB mutant K476C, bound to casein, post-state | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Alpha-S1-casein, Hyperactive disaggregase ClpB, ... | | Authors: | Rizo, A.R, Lin, J.-B, Gates, S.N, Tse, E, Bart, S.M, Castellano, L.M, Dimaio, F, Shorter, J, Southworth, D.R. | | Deposit date: | 2019-03-18 | | Release date: | 2019-06-12 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural basis for substrate gripping and translocation by the ClpB AAA+ disaggregase.
Nat Commun, 10, 2019
|
|
6OG3
 
 | | Focus classification structure of the hyperactive ClpB mutant K476C, bound to casein, NTD-trimer | | Descriptor: | Alpha S1-casein, Hyperactive disaggregase ClpB | | Authors: | Rizo, A.R, Lin, J.-B, Gates, S.N, Tse, E, Bart, S.M, Castellano, L.M, Dimaio, F, Shorter, J, Southworth, D.R. | | Deposit date: | 2019-04-01 | | Release date: | 2019-06-12 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Structural basis for substrate gripping and translocation by the ClpB AAA+ disaggregase.
Nat Commun, 10, 2019
|
|
5E8D
 
 | | Crystal structure of human epiregulin in complex with the Fab fragment of murine monoclonal antibody 9E5 | | Descriptor: | CHLORIDE ION, GLYCEROL, Proepiregulin, ... | | Authors: | Kado, Y, Mizohata, E, Nagatoishi, S, Iijima, M, Shinoda, K, Miyafusa, T, Nakayama, T, Yoshizumi, T, Sugiyama, A, Kawamura, T, Lee, Y.H, Matsumura, H, Doi, H, Fujitani, H, Kodama, T, Shibasaki, Y, Tsumoto, K, Inoue, T. | | Deposit date: | 2015-10-14 | | Release date: | 2015-12-09 | | Last modified: | 2020-02-19 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Epiregulin Recognition Mechanisms by Anti-epiregulin Antibody 9E5: STRUCTURAL, FUNCTIONAL, AND MOLECULAR DYNAMICS SIMULATION ANALYSES
J.Biol.Chem., 291, 2016
|
|
5CI9
 
 | | Crystal structure of human Tob in complex with inhibitor fragment 6 | | Descriptor: | 1-(propan-2-yl)-1H-benzimidazole-5-carboxylic acid, Protein Tob1, SODIUM ION | | Authors: | Bai, Y, Tashiro, S, Nagatoishi, S, Suzuki, T, Tsumoto, K, Bartlam, M, Yamamoto, T. | | Deposit date: | 2015-07-11 | | Release date: | 2015-11-18 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis for inhibition of the Tob-CNOT7 interaction by a fragment screening approach
Protein Cell, 6, 2015
|
|
5CI8
 
 | | Crystal structure of human Tob in complex with inhibitor fragment 1 | | Descriptor: | Protein Tob1, pyrrolo[1,2-a]quinoxalin-4(5H)-one | | Authors: | Bai, Y, Tashiro, S, Nagatoishi, S, Suzuki, T, Tsumoto, K, Bartlam, M, Yamamoto, T. | | Deposit date: | 2015-07-11 | | Release date: | 2015-11-18 | | Last modified: | 2015-12-09 | | Method: | X-RAY DIFFRACTION (2.328 Å) | | Cite: | Structural basis for inhibition of the Tob-CNOT7 interaction by a fragment screening approach
Protein Cell, 6, 2015
|
|
3F06
 
 | | Crystal Structure Analysis of Human HDAC8 D101A Variant. | | Descriptor: | 4-(dimethylamino)-N-[7-(hydroxyamino)-7-oxoheptyl]benzamide, BETA-MERCAPTOETHANOL, Histone deacetylase 8, ... | | Authors: | Dowling, D.P, Gantt, S.L, Gattis, S.G, Fierke, C.A, Christianson, D.W. | | Deposit date: | 2008-10-24 | | Release date: | 2008-12-30 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structural studies of human histone deacetylase 8 and its site-specific variants complexed with substrate and inhibitors.
Biochemistry, 47, 2008
|
|
3EZT
 
 | | Crystal Structure Analysis of Human HDAC8 D101E Variant | | Descriptor: | 4-(dimethylamino)-N-[7-(hydroxyamino)-7-oxoheptyl]benzamide, BETA-MERCAPTOETHANOL, Histone deacetylase 8, ... | | Authors: | Dowling, D.P, Gantt, S.L, Gattis, S.G, Fierke, C.A, Christianson, D.W. | | Deposit date: | 2008-10-23 | | Release date: | 2008-12-30 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Structural studies of human histone deacetylase 8 and its site-specific variants complexed with substrate and inhibitors.
Biochemistry, 47, 2008
|
|
3EZP
 
 | | Crystal Structure Analysis of human HDAC8 D101N variant | | Descriptor: | 4-(dimethylamino)-N-[7-(hydroxyamino)-7-oxoheptyl]benzamide, BETA-MERCAPTOETHANOL, Histone deacetylase 8, ... | | Authors: | Dowling, D.P, Gantt, S.L, Gattis, S.G, Fierke, C.A, Christianson, D.W. | | Deposit date: | 2008-10-23 | | Release date: | 2008-12-30 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structural studies of human histone deacetylase 8 and its site-specific variants complexed with substrate and inhibitors.
Biochemistry, 47, 2008
|
|
3MZ6
 
 | | Crystal structure of D101L Fe2+ HDAC8 complexed with M344 | | Descriptor: | 4-(dimethylamino)-N-[7-(hydroxyamino)-7-oxoheptyl]benzamide, FE (II) ION, Histone deacetylase 8, ... | | Authors: | Dowling, D.P, Gattis, S.G, Fierke, C.A, Christianson, D.W. | | Deposit date: | 2010-05-11 | | Release date: | 2010-06-23 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structures of metal-substituted human histone deacetylase 8 provide mechanistic inferences on biological function.
Biochemistry, 49, 2010
|
|
3MZ7
 
 | | Crystal structure of D101L Co2+ HDAC8 complexed with M344 | | Descriptor: | 4-(dimethylamino)-N-[7-(hydroxyamino)-7-oxoheptyl]benzamide, COBALT (II) ION, GLYCEROL, ... | | Authors: | Dowling, D.P, Gattis, S.G, Fierke, C.A, Christianson, D.W. | | Deposit date: | 2010-05-11 | | Release date: | 2010-06-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structures of metal-substituted human histone deacetylase 8 provide mechanistic inferences on biological function.
Biochemistry, 49, 2010
|
|
