8T77
 
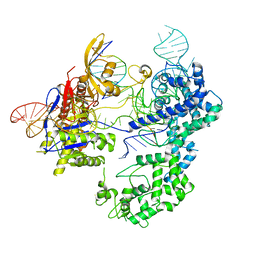 | | SpRY-Cas9:gRNA complex bound to non-target DNA with 6 bp R-loop | | Descriptor: | CRISPR-associated endonuclease Cas9/Csn1, NTS, TS, ... | | Authors: | Hibshman, G.N, Bravo, J.P.K, Taylor, D.W. | | Deposit date: | 2023-06-19 | | Release date: | 2024-05-01 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.25 Å) | | Cite: | Unraveling the mechanisms of PAMless DNA interrogation by SpRY-Cas9.
Nat Commun, 15, 2024
|
|
8T6X
 
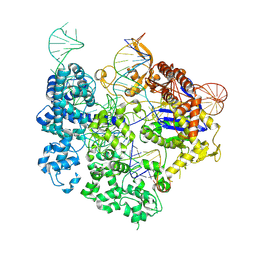 | | SpRY-Cas9:gRNA complex targeting TAC PAM DNA with 18 bp R-loop | | Descriptor: | CRISPR-associated endonuclease Cas9/Csn1, MAGNESIUM ION, NTS, ... | | Authors: | Hibshman, G.N, Bravo, J.P.K, Taylor, D.W. | | Deposit date: | 2023-06-19 | | Release date: | 2024-05-01 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.08 Å) | | Cite: | Unraveling the mechanisms of PAMless DNA interrogation by SpRY-Cas9.
Nat Commun, 15, 2024
|
|
8T79
 
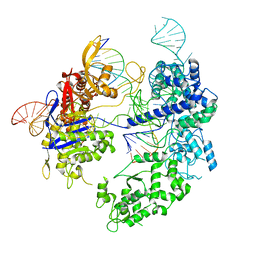 | | SpRY-Cas9:gRNA complex bound to non-target DNA with 10 bp R-loop | | Descriptor: | CRISPR-associated endonuclease Cas9/Csn1, NTS, TS, ... | | Authors: | Hibshman, G.N, Bravo, J.P.K, Taylor, D.W. | | Deposit date: | 2023-06-20 | | Release date: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (3.04 Å) | | Cite: | Unraveling the mechanisms of PAMless DNA interrogation by SpRY-Cas9.
Nat Commun, 15, 2024
|
|
6DA7
 
 | | Crystal structure of the TtnD decarboxylase from the tautomycetin biosynthesis pathway of Streptomyces griseochromogenes with apo form at 1.83 A resolution (I222) | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, GLYCEROL, SODIUM ION, ... | | Authors: | Han, L, Rudolf, J.D, Chang, C.-Y, Miller, M.D, Soman, J, Phillips Jr, G.N, Shen, B, Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2018-05-01 | | Release date: | 2018-10-03 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Biochemical and Structural Characterization of TtnD, a Prenylated FMN-Dependent Decarboxylase from the Tautomycetin Biosynthetic Pathway.
ACS Chem. Biol., 13, 2018
|
|
6DRU
 
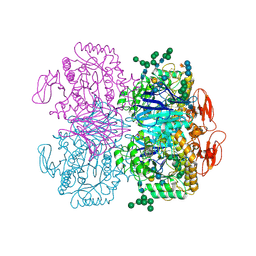 | | Xylosidase from Aspergillus niger | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, Glycosyl hydrolases family 31 family protein, ... | | Authors: | Cao, H, Xu, W, Betancourt, M, Walton, J.D, Brumm, P, Phillips Jr, G.N, Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2018-06-13 | | Release date: | 2018-08-22 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal Structure of alpha-Xylosidase fromAspergillus nigerin Complex with a Hydrolyzed Xyloglucan Product and New Insights in Accurately Predicting Substrate Specificities of GH31 Family Glycosidases.
Acs Sustain Chem Eng, 8, 2020
|
|
6D2V
 
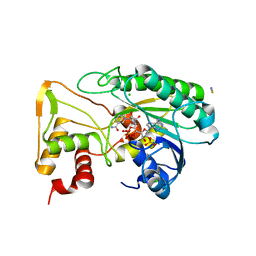 | | Apo Structure of TerB, an NADP Dependent Oxidoreductase in the Terfestatin Biosynthesis Pathway | | Descriptor: | CHLORIDE ION, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, THIOCYANATE ION, ... | | Authors: | Clinger, J.A, Elshahawi, S.I, Zhang, Y, Hall, R.P, Liu, Y, Thorson, J.S, Phillips Jr, G.N. | | Deposit date: | 2018-04-14 | | Release date: | 2018-06-13 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.901 Å) | | Cite: | Structure and Function of Terfestatin Biosynthesis Enzymes TerB and TerC
To Be Published
|
|
6D34
 
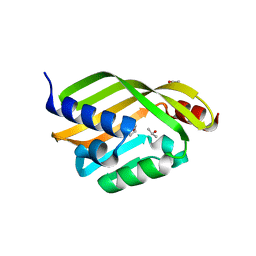 | | Apo Crystal Structure of TerC, a Terfestatin Biosynthesis Enzyme | | Descriptor: | ISOPROPYL ALCOHOL, TerC | | Authors: | Clinger, J.A, Elshahawi, S.I, Zhang, Y, Hall, R.P, Liu, Y, Thorson, J.S, Phillips Jr, G.N. | | Deposit date: | 2018-04-14 | | Release date: | 2018-06-13 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure and Function of Terfestatin Biosynthesis Enzymes TerB and TerC
To Be Published
|
|
4OPF
 
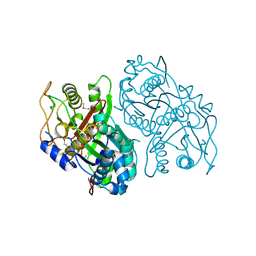 | | Streptomcyes albus JA3453 oxazolomycin ketosynthase domain OzmH KS8 | | Descriptor: | NRPS/PKS | | Authors: | Osipiuk, J, Bigelow, L, Endres, M, Babnigg, G, Bingman, C.A, Yennamalli, R, Lohman, J.R, Ma, M, Shen, B, Phillips Jr, G.N, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2014-02-05 | | Release date: | 2014-02-19 | | Last modified: | 2017-11-22 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Structural and evolutionary relationships of "AT-less" type I polyketide synthase ketosynthases.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
6DA9
 
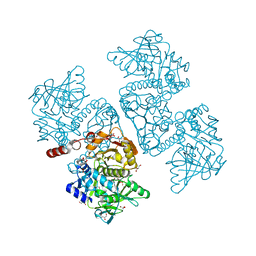 | | Crystal structure of the TtnD decarboxylase from the tautomycetin biosynthesis pathway of Streptomyces griseochromogenes with FMN bound at 2.05 A resolution | | Descriptor: | FLAVIN MONONUCLEOTIDE, GLYCEROL, MANGANESE (II) ION, ... | | Authors: | Han, L, Rudolf, J.D, Chang, C.-Y, Miller, M.D, Soman, J, Xu, W, Phillips Jr, G.N, Shen, B, Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2018-05-01 | | Release date: | 2018-10-03 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Biochemical and Structural Characterization of TtnD, a Prenylated FMN-Dependent Decarboxylase from the Tautomycetin Biosynthetic Pathway.
ACS Chem. Biol., 13, 2018
|
|
4Q29
 
 | | Ensemble Refinement of plu4264 protein from Photorhabdus luminescens | | Descriptor: | NICKEL (II) ION, SODIUM ION, plu4264 protein | | Authors: | Wang, F, Michalska, K, Li, H, Jedrzejczak, R, Babnigg, G, Bingman, C.A, Yennamalli, R, Weerth, S, Miller, M.D, Thomas, M.G, Joachimiak, A, Phillips Jr, G.N, Enzyme Discovery for Natural Product Biosynthesis (NatPro), Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2014-04-07 | | Release date: | 2014-05-07 | | Last modified: | 2015-02-11 | | Method: | X-RAY DIFFRACTION (1.349 Å) | | Cite: | Structure of a cupin protein Plu4264 from Photorhabdus luminescens subsp. laumondii TTO1 at 1.35 angstrom resolution.
Proteins, 83, 2015
|
|
8TIB
 
 | | Cryo-EM of tri-pilus from S. islandicus REY15A | | Descriptor: | DUF973 family protein | | Authors: | Eastep, G.N, Liu, J, Rich-New, S.T, Egelman, E.H, Krupovic, M, Wang, F. | | Deposit date: | 2023-07-19 | | Release date: | 2024-01-10 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.47 Å) | | Cite: | Two distinct archaeal type IV pili structures formed by proteins with identical sequence.
Nat Commun, 15, 2024
|
|
8TIF
 
 | | Cryo-EM of mono-pilus from S. islandicus REY15A | | Descriptor: | DUF973 family protein | | Authors: | Eastep, G.N, Liu, J, Rich-New, S.T, Egelman, E.H, Krupovic, M, Wang, F. | | Deposit date: | 2023-07-19 | | Release date: | 2024-01-10 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.89 Å) | | Cite: | Two distinct archaeal type IV pili structures formed by proteins with identical sequence.
Nat Commun, 15, 2024
|
|
4PYT
 
 | | Crystal structure of a MurB family EP-UDP-N-acetylglucosamine reductase | | Descriptor: | CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, MAGNESIUM ION, ... | | Authors: | Cao, H, Franz, L, Sen, S, Bingman, C.A, Auldridge, M, Steinmetz, E, Mead, D, Phillips Jr, G.N. | | Deposit date: | 2014-03-27 | | Release date: | 2014-05-21 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.853 Å) | | Cite: | LucY: A Versatile New Fluorescent Reporter Protein.
Plos One, 10, 2015
|
|
4MV2
 
 | | Crystal structure of plu4264 protein from Photorhabdus luminescens | | Descriptor: | NICKEL (II) ION, SODIUM ION, plu4264 | | Authors: | Michalska, K, Li, H, Jedrzejczak, R, Babnigg, G, Bingman, C.A, Yennamalli, R, Weerth, S, Thomas, M.G, Phillips Jr, G.N, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2013-09-23 | | Release date: | 2013-10-02 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.349 Å) | | Cite: | Structure of a cupin protein Plu4264 from Photorhabdus luminescens subsp. laumondii TTO1 at 1.35 angstrom resolution.
Proteins, 83, 2015
|
|
4Q31
 
 | | The crystal structure of cystathione gamma lyase (CalE6) from Micromonospora echinospora | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CHLORIDE ION, FORMIC ACID, ... | | Authors: | Tan, K, Bigelow, L, Jedrzejczak, R, Babnigg, G, Bingman, C.A, Yennamalli, R.M, Singh, S, Kharel, M.K, Thorson, J.S, Phillips Jr, G.N, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2014-04-10 | | Release date: | 2014-05-07 | | Last modified: | 2017-03-08 | | Method: | X-RAY DIFFRACTION (2.099 Å) | | Cite: | Structural dynamics of a methionine gamma-lyase for calicheamicin biosynthesis: Rotation of the conserved tyrosine stacking with pyridoxal phosphate.
Struct Dyn, 3, 2016
|
|
3TSR
 
 | | X-ray structure of mouse ribonuclease inhibitor complexed with mouse ribonuclease 1 | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, Ribonuclease inhibitor, ... | | Authors: | Chang, A, Lomax, J.E, Bingman, C.A, Raines, R.T, Phillips Jr, G.N. | | Deposit date: | 2011-09-13 | | Release date: | 2012-09-19 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.1999 Å) | | Cite: | Functional evolution of ribonuclease inhibitor: insights from birds and reptiles.
J.Mol.Biol., 426, 2014
|
|
4OQJ
 
 | | Streptomcyes albus JA3453 oxazolomycin ketosynthase domain OzmQ KS1 | | Descriptor: | GLYCEROL, PHOSPHATE ION, PKS, ... | | Authors: | Nocek, B, Mack, J, Endras, M, Babnigg, G, Bingman, C.A, Yennamalli, R, Lohman, J.R, Ma, M, Shen, B, Phillips Jr, G.N, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2014-02-09 | | Release date: | 2014-03-19 | | Last modified: | 2016-11-02 | | Method: | X-RAY DIFFRACTION (1.904 Å) | | Cite: | Structural and evolutionary relationships of "AT-less" type I polyketide synthase ketosynthases.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
4R82
 
 | | Streptomyces globisporus C-1027 NADH:FAD oxidoreductase SgcE6 in complex with NAD and FAD fragments | | Descriptor: | ACETATE ION, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Tan, K, Bigelow, L, Clancy, S, Babnigg, G, Bingman, C.A, Yennamalli, R, Lohman, J.R, Ma, M, Shen, B, Phillips Jr, G.N, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2014-08-29 | | Release date: | 2014-10-01 | | Last modified: | 2016-11-02 | | Method: | X-RAY DIFFRACTION (1.659 Å) | | Cite: | Crystal Structures of SgcE6 and SgcC, the Two-Component Monooxygenase That Catalyzes Hydroxylation of a Carrier Protein-Tethered Substrate during the Biosynthesis of the Enediyne Antitumor Antibiotic C-1027 in Streptomyces globisporus.
Biochemistry, 55, 2016
|
|
4QYR
 
 | | Streptomyces platensis isomigrastatin ketosynthase domain MgsE KS3 | | Descriptor: | ACETIC ACID, AT-less polyketide synthase, CHLORIDE ION, ... | | Authors: | Kim, Y, Li, H, Endres, M, Babnigg, J, Bingman, C.A, Yennamalli, R, Lohman, J.R, Ma, M, Shen, B, Phillips Jr, G.N, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2014-07-25 | | Release date: | 2014-08-20 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.902 Å) | | Cite: | Structural and evolutionary relationships of "AT-less" type I polyketide synthase ketosynthases.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
6WF4
 
 | | Crystal Structure of TerC Co-crystallized with Polyporic Acid | | Descriptor: | (2~5~S)-2~3~,2~5~,2~6~-trihydroxy[1~1~,2~1~:2~4~,3~1~-terphenyl]-2~2~(2~5~H)-one, ISOPROPYL ALCOHOL, Terfestatin Biosyntheis Enzyme C | | Authors: | Clinger, J.A, Miller, M.D, Hall, R.E, Zhang, Y, Elshahawi, S.I, Thorson, J.S, Van Lanen, S.G, Phillips Jr, G.N, Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2020-04-03 | | Release date: | 2021-04-07 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Structural and functional characterization of two cooperative enzymes responsible for the stability
of p-terphenyls.
To be published
|
|
6WFV
 
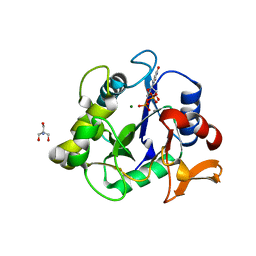 | | The crystal structure of a collagen galactosylhydroxylysyl glucosyltransferase from human | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, MAGNESIUM ION, MANGANESE (II) ION, ... | | Authors: | Guo, H.-F, Tsai, C.-L, Miller, M.D, Phillips Jr, G.N, Tainer, J.A, Kurie, J.M. | | Deposit date: | 2020-04-04 | | Release date: | 2021-04-14 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The crystal structure of a collagen galactosylhydroxylysyl glucosyltransferase from human
To Be Published
|
|
6XN8
 
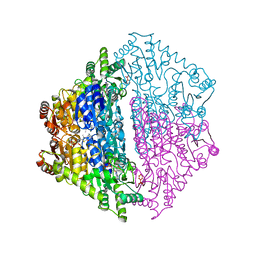 | | Crystal Structure of 2-hydroxyacyl CoA lyase (HACL) from Rhodospirillales bacterium URHD0017 | | Descriptor: | 2-hydroxyacyl-CoA lyase 1, 2-{3-[(4-AMINO-2-METHYLPYRIMIDIN-5-YL)METHYL]-4-METHYL-2-OXO-2,3-DIHYDRO-1,3-THIAZOL-5-YL}ETHYL TRIHYDROGEN DIPHOSPHATE, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Miller, M.D, Xu, W, Olmos Jr, J.L, Chou, A, Clomburg, J.M, Gonzalez, R, Philips Jr, G.N. | | Deposit date: | 2020-07-02 | | Release date: | 2021-07-07 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal Structure of 2-hydroxyacyl CoA lyase (HACL) from Rhodospirillales bacterium URHD0017
To Be Published
|
|
6XRQ
 
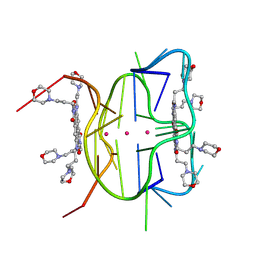 | | Structural descriptions of ligand interactions to DNA and RNA quadruplexes folded from the non-coding region of Pseudorabies virus | | Descriptor: | 2,7-bis[3-(morpholin-4-yl)propyl]-4,9-bis{[3-(morpholin-4-yl)propyl]amino}benzo[lmn][3,8]phenanthroline-1,3,6,8(2H,7H)-tetrone, POTASSIUM ION, RNA (5' GP*GP*CP*UP*CP*GP*GP*CP*GP*GP*CP*GP*GP*A-3') | | Authors: | Zhang, Y.S, Parkinson, G.N, Wei, D.G. | | Deposit date: | 2020-07-13 | | Release date: | 2021-07-28 | | Last modified: | 2024-06-26 | | Method: | X-RAY DIFFRACTION (1.21 Å) | | Cite: | Structural descriptions of ligand interactions to RNA quadruplexes folded from the non-coding region of Pseudorabies virus.
Biochimie, 2024
|
|
1SPE
 
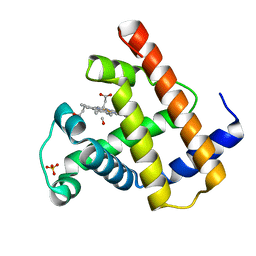 | | SPERM WHALE NATIVE CO MYOGLOBIN AT PH 4.0, TEMP 4C | | Descriptor: | CARBON MONOXIDE, MYOGLOBIN, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Yang, F, Phillips Jr, G.N. | | Deposit date: | 1995-10-25 | | Release date: | 1996-03-08 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structures of CO-, deoxy- and met-myoglobins at various pH values.
J.Mol.Biol., 256, 1996
|
|
3BVO
 
 | | Crystal structure of human co-chaperone protein HscB | | Descriptor: | Co-chaperone protein HscB, mitochondrial precursor, SULFATE ION, ... | | Authors: | Bitto, E, Bingman, C.A, McCoy, J.G, Wesenberg, G.E, Phillips Jr, G.N, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2008-01-07 | | Release date: | 2008-01-15 | | Last modified: | 2017-10-25 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure of human J-type co-chaperone HscB reveals a tetracysteine metal-binding domain.
J.Biol.Chem., 283, 2008
|
|
