2QIO
 
 | |
3LBJ
 
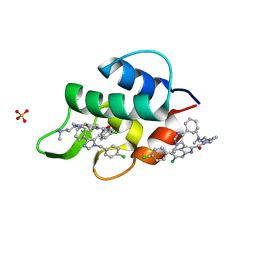 | | Structure of human MDMX protein in complex with a small molecule inhibitor | | Descriptor: | N-[(3S)-1-({6-chloro-3-[1-(4-chlorobenzyl)-4-phenyl-1H-imidazol-5-yl]-1H-indol-2-yl}carbonyl)pyrrolidin-3-yl]-N,N',N'-trimethylpropane-1,3-diamine, Protein Mdm4, SULFATE ION | | Authors: | Popowicz, G.M, Czarna, A, Wolf, S, Holak, T.A. | | Deposit date: | 2010-01-08 | | Release date: | 2010-03-16 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structures of low molecular weight inhibitors bound to MDMX and MDM2 reveal new approaches for p53-MDMX/MDM2 antagonist drug discovery
Cell Cycle, 9, 2010
|
|
2QS1
 
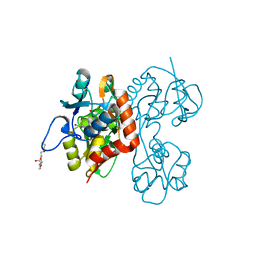 | | Crystal structure of the GluR5 ligand binding core dimer in complex with UBP315 at 1.80 Angstroms resolution | | Descriptor: | 3-({3-[(2S)-2-amino-2-carboxyethyl]-5-methyl-2,6-dioxo-3,6-dihydropyrimidin-1(2H)-yl}methyl)-4,5-dibromothiophene-2-carboxylic acid, CHLORIDE ION, Glutamate receptor, ... | | Authors: | Alushin, G.M, Jane, D.E, Mayer, M.L. | | Deposit date: | 2007-07-30 | | Release date: | 2008-08-05 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Binding site and ligand flexibility revealed by high resolution crystal structures of GluK1 competitive antagonists.
Neuropharmacology, 60, 2011
|
|
2P9T
 
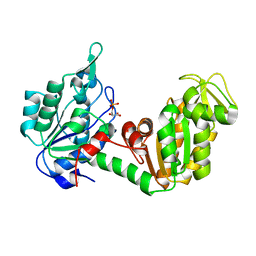 | | Crystal Structure of Phosphoglycerate Kinase-2 bound to 3-phosphoglycerate | | Descriptor: | 3-PHOSPHOGLYCERIC ACID, Phosphoglycerate kinase, testis specific | | Authors: | Sawyer, G.M, Monzingo, A.F, Poteet, E.C, Robertus, J.D. | | Deposit date: | 2007-03-26 | | Release date: | 2007-11-27 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | X-ray analysis of phosphoglycerate kinase 2, a sperm-specific isoform from Mus musculus.
Proteins, 71, 2007
|
|
2QS3
 
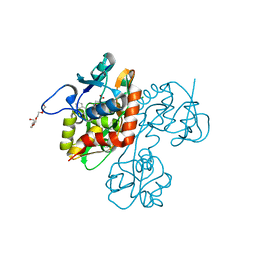 | | Crystal structure of the GluR5 ligand binding core dimer in complex with UBP316 at 1.76 Angstroms resolution | | Descriptor: | 3-({3-[(2S)-2-amino-2-carboxyethyl]-5-methyl-2,6-dioxo-3,6-dihydropyrimidin-1(2H)-yl}methyl)-5-phenylthiophene-2-carboxylic acid, CHLORIDE ION, Glutamate receptor, ... | | Authors: | Alushin, G.M, Jane, D.E, Mayer, M.L. | | Deposit date: | 2007-07-30 | | Release date: | 2008-08-05 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | ACET is a highly potent and specific kainate receptor antagonist: characterisation and effects on hippocampal mossy fibre function.
Neuropharmacology, 56, 2009
|
|
2QE7
 
 | | Crystal structure of the f1-atpase from the thermoalkaliphilic bacterium bacillus sp. ta2.a1 | | Descriptor: | ATP synthase subunit alpha, ATP synthase subunit beta, ATP synthase subunit epsilon, ... | | Authors: | Stocker, A, Keis, S, Vonck, J, Cook, G.M, Dimroth, P. | | Deposit date: | 2007-06-25 | | Release date: | 2007-08-21 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.06 Å) | | Cite: | The Structural Basis for Unidirectional Rotation of Thermoalkaliphilic F(1)-ATPase.
Structure, 15, 2007
|
|
2QS2
 
 | | Crystal structure of the GluR5 ligand binding core dimer in complex with UBP318 at 1.80 Angstroms resolution | | Descriptor: | 3-({3-[(2S)-2-amino-2-carboxyethyl]-5-bromo-2,6-dioxo-3,6-dihydropyrimidin-1(2H)-yl}methyl)thiophene-2-carboxylic acid, CHLORIDE ION, Glutamate receptor, ... | | Authors: | Alushin, G.M, Jane, D.E, Mayer, M.L. | | Deposit date: | 2007-07-30 | | Release date: | 2008-08-05 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Binding site and ligand flexibility revealed by high resolution crystal structures of GluK1 competitive antagonists.
Neuropharmacology, 60, 2011
|
|
1VTR
 
 | | STRUCTURE OF THE DEOXYTETRANUCLEOTIDE D-PAPTPAPT AND A SEQUENCE-DEPENDENT MODEL FOR POLY(DA-DT) | | Descriptor: | DNA (5'-D(*AP*TP*AP*T)-3') | | Authors: | Viswamitra, M.A, Shakked, Z, Jones, P.G, Sheldrick, G.M, Salisbury, S.A, Kennard, O. | | Deposit date: | 1988-08-18 | | Release date: | 2011-07-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.04 Å) | | Cite: | Structure of the Deoxytetranucleotide d-pApTpApT and a Sequence-Dependent Model for Poly(dA-dT)
Biopolymers, 21, 1982
|
|
1WJF
 
 | | SOLUTION STRUCTURE OF H12C MUTANT OF THE N-TERMINAL ZN BINDING DOMAIN OF HIV-1 INTEGRASE COMPLEXED TO CADMIUM, NMR, 40 STRUCTURES | | Descriptor: | CADMIUM ION, HIV-1 INTEGRASE | | Authors: | Cai, M, Gronenborn, A.M, Clore, G.M. | | Deposit date: | 1998-06-11 | | Release date: | 1998-12-16 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the His12 --> Cys mutant of the N-terminal zinc binding domain of HIV-1 integrase complexed to cadmium.
Protein Sci., 7, 1998
|
|
5U06
 
 | | Grb7-SH2 with bicyclic peptide inhibitor containing a pY mimetic | | Descriptor: | Growth factor receptor-bound protein 7, POTASSIUM ION, bicyclic peptide inhibitor: LYS-PHE-GLU-GLY-CMF-ASP-ASN-GLU-CST | | Authors: | Watson, G.M, Wilce, J.A. | | Deposit date: | 2016-11-22 | | Release date: | 2017-11-15 | | Last modified: | 2020-01-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Discovery, Development, and Cellular Delivery of Potent and Selective Bicyclic Peptide Inhibitors of Grb7 Cancer Target.
J. Med. Chem., 60, 2017
|
|
5TWV
 
 | | Cryo-EM structure of the pancreatic ATP-sensitive K+ channel SUR1/Kir6.2 in the presence of ATP and glibenclamide | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, ATP-binding cassette sub-family C member 8, ATP-sensitive inward rectifier potassium channel 11 | | Authors: | Martin, G.M, Yoshioka, C, Chen, J.Z, Shyng, S.L. | | Deposit date: | 2016-11-14 | | Release date: | 2017-01-25 | | Last modified: | 2019-12-25 | | Method: | ELECTRON MICROSCOPY (6.3 Å) | | Cite: | Cryo-EM structure of the ATP-sensitive potassium channel illuminates mechanisms of assembly and gating.
Elife, 6, 2017
|
|
5U1Q
 
 | | Grb7-SH2 with bicyclic peptide inhibitor | | Descriptor: | CHLORIDE ION, Growth factor receptor-bound protein 7, LYS-PHE-GLU-GLY-TYR-ASP-ASN-GLU-CST | | Authors: | Watson, G.M, Wilce, M.C.J, Wilce, J.A. | | Deposit date: | 2016-11-28 | | Release date: | 2017-11-15 | | Last modified: | 2020-01-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Discovery, Development, and Cellular Delivery of Potent and Selective Bicyclic Peptide Inhibitors of Grb7 Cancer Target.
J. Med. Chem., 60, 2017
|
|
5TYI
 
 | | Grb7 SH2 with bicyclic peptide containing pY mimetic | | Descriptor: | Growth factor receptor-bound protein 7, Peptide inhibitor | | Authors: | Watson, G.M, Wilce, M.C.J, Wilce, J.A. | | Deposit date: | 2016-11-20 | | Release date: | 2017-11-15 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Discovery, Development, and Cellular Delivery of Potent and Selective Bicyclic Peptide Inhibitors of Grb7 Cancer Target.
J. Med. Chem., 60, 2017
|
|
1FHI
 
 | | SUBSTRATE ANALOG (IB2) COMPLEX WITH THE FRAGILE HISTIDINE TRIAD PROTEIN, FHIT | | Descriptor: | FRAGILE HISTIDINE TRIAD PROTEIN, P1-P2-METHYLENE-P3-THIO-DIADENOSINE TRIPHOSPHATE | | Authors: | Pace, H.C, Garrison, P.N, Barnes, L.D, Draganescu, A, Rosler, A, Blackburn, G.M, Siprashvili, Z, Croce, C.M, Huebner, K, Brenner, C. | | Deposit date: | 1997-12-11 | | Release date: | 1998-06-17 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Genetic, biochemical, and crystallographic characterization of Fhit-substrate complexes as the active signaling form of Fhit.
Proc.Natl.Acad.Sci.USA, 95, 1998
|
|
1GJJ
 
 | |
1GIP
 
 | |
1GYO
 
 | | Crystal structure of the di-tetraheme cytochrome c3 from Desulfovibrio gigas at 1.2 Angstrom resolution | | Descriptor: | CYTOCHROME C3, A DIMERIC CLASS III C-TYPE CYTOCHROME, GLYCEROL, ... | | Authors: | Aragao, D, Frazao, C, Sieker, L, Sheldrick, G.M, Legall, J, Carrondo, M.A. | | Deposit date: | 2002-04-29 | | Release date: | 2002-05-24 | | Last modified: | 2023-03-29 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Structure of Dimeric Cytochrome C3 from Desulfovibrio Gigas at 1.2 A Resolution
Acta Crystallogr.,Sect.D, 59, 2003
|
|
1HUN
 
 | |
1HUM
 
 | |
2W7S
 
 | | SplA serine protease of Staphylococcus aureus (1.8A) | | Descriptor: | SERINE PROTEASE SPLA | | Authors: | Stec-Niemczyka, J, Pustelny, K, Kisielewska, M, Bista, M, Boulware, K.T, Stennicke, H.R, Thogersen, I.B, Daugherty, P.S, Enghild, J.J, Popowicz, G.M, Dubin, A, Potempa, J, Dubin, G. | | Deposit date: | 2008-12-30 | | Release date: | 2010-03-31 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural and Functional Characterization of Spla, an Exclusively Specific Protease of Staphylococcus Aureus
Biochem.J., 419, 2009
|
|
2VID
 
 | | Serine protease SplB from Staphylococcus aureus at 1.8A resolution | | Descriptor: | SERINE PROTEASE SPLB | | Authors: | Dubin, G, Stec-Niemczyk, J, Kisielewska, M, Pustelny, K, Popowicz, G.M, Bista, M, Kantyka, T, Boulware, K.T, Stennicke, H.R, Czarna, A, Phopaisarn, M, Daugherty, P.S, Thogersen, I.B, Enghild, J.J, Thornberry, N, Dubin, A, Potempa, J. | | Deposit date: | 2007-11-30 | | Release date: | 2008-05-13 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Enzymatic Activity of the Staphylococcus Aureus Splb Serine Protease is Induced by Substrates Containing the Sequence Trp-Glu-Leu-Gln.
J.Mol.Biol., 379, 2008
|
|
2WY2
 
 | | NMR structure of the IIAchitobiose-IIBchitobiose phosphoryl transition state complex of the N,N'-diacetylchitoboise brance of the E. coli phosphotransferase system. | | Descriptor: | N,N'-DIACETYLCHITOBIOSE-SPECIFIC PHOSPHOTRANSFERASE ENZYME IIA COMPONENT, N,N'-DIACETYLCHITOBIOSE-SPECIFIC PHOSPHOTRANSFERASE ENZYME IIB COMPONENT, PHOSPHITE ION | | Authors: | Sang, Y.S, Cai, M, Clore, G.M. | | Deposit date: | 2009-11-11 | | Release date: | 2009-12-08 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of the Iiachitobose-Iibchitobiose Complex of the N,N'-Diacetylchitobiose Branch of the Escherichia Coli Phosphotransfer System
J.Biol.Chem., 285, 2010
|
|
2W7U
 
 | | SplA serine protease of Staphylococcus aureus (2.4A) | | Descriptor: | SERINE PROTEASE SPLA | | Authors: | Stec-Niemczyka, J, Pustelny, K, Kisielewska, M, Bista, M, Boulware, K.T, Stennicke, H.R, Thogersen, I.B, Daugherty, P.S, Enghild, J.J, Popowicz, G.M, Dubin, A, Potempa, J, Dubin, G. | | Deposit date: | 2008-12-30 | | Release date: | 2010-03-31 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Structural and Functional Characterization of Spla, an Exclusively Specific Protease of Staphylococcus Aureus.
Biochem.J., 419, 2009
|
|
2WWV
 
 | | NMR structure of the IIAchitobiose-IIBchitobiose complex of the N,N'- diacetylchitoboise brance of the E. coli phosphotransferase system. | | Descriptor: | N,N'-DIACETYLCHITOBIOSE-SPECIFIC PHOSPHOTRANSFERASE ENZYME IIA COMPONENT, N,N'-DIACETYLCHITOBIOSE-SPECIFIC PHOSPHOTRANSFERASE ENZYME IIB COMPONENT | | Authors: | Sang, Y.S, Cai, M, Clore, G.M. | | Deposit date: | 2009-10-29 | | Release date: | 2009-12-08 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of the Iiachitobose-Iibchitobiose Complex of the N,N'-Diacetylchitobiose Branch of the Escherichia Coli Phosphotransfer System
J.Biol.Chem., 285, 2010
|
|
6R78
 
 | | Structure of IMP-13 metallo-beta-lactamase in apo form (loop closed) | | Descriptor: | 1,2-ETHANEDIOL, BETA-MERCAPTOETHANOL, Beta-lactamase, ... | | Authors: | Zak, K.M, Softley, C, Kolonko, M, Sattler, M, Popowicz, G.M. | | Deposit date: | 2019-03-28 | | Release date: | 2020-04-01 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Structure and Molecular Recognition Mechanism of IMP-13 Metallo-beta-Lactamase.
Antimicrob.Agents Chemother., 64, 2020
|
|
