8G2K
 
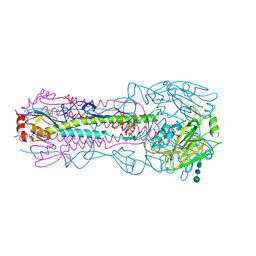 | |
7P49
 
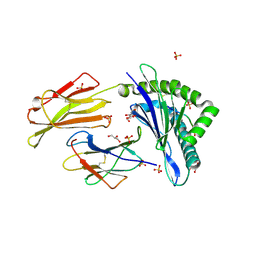 | | HLA-E*01:03 in complex with Mtb14 | | Descriptor: | Beta-2-microglobulin, GLYCEROL, HLA class I histocompatibility antigen, ... | | Authors: | Walters, L.C, Gillespie, G.M. | | Deposit date: | 2021-07-10 | | Release date: | 2022-06-22 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Primary and secondary functions of HLA-E are determined by stability and conformation of the peptide-bound complexes.
Cell Rep, 39, 2022
|
|
7P4B
 
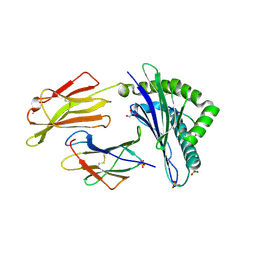 | | HLA-E*01:03 in complex with IL9 | | Descriptor: | Beta-2-microglobulin, ESAT-6-like protein EsxH, GLYCEROL, ... | | Authors: | Walters, L.C, Gillespie, G.M. | | Deposit date: | 2021-07-10 | | Release date: | 2022-07-27 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Primary and secondary functions of HLA-E are determined by stability and conformation of the peptide-bound complexes.
Cell Rep, 39, 2022
|
|
4YGK
 
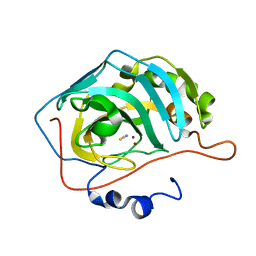 | | NaSCN--Interactions between Hofmeister Anions and the Binding Pocket of a Protein | | Descriptor: | Carbonic anhydrase 2, HYDROXIDE ION, THIOCYANATE ION, ... | | Authors: | Fox, J.M, Kang, K, Sherman, W, Heroux, A, Sastry, G.M, Baghbanzadeh, M, Lockett, M.R, Whitesides, G.M. | | Deposit date: | 2015-02-26 | | Release date: | 2015-03-25 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Interactions between Hofmeister Anions and the Binding Pocket of a Protein.
J.Am.Chem.Soc., 137, 2015
|
|
4YGJ
 
 | | NaBr--Interactions between Hofmeister Anions and the Binding Pocket of a Protein | | Descriptor: | BROMIDE ION, Carbonic anhydrase 2, ZINC ION | | Authors: | Fox, J.M, Kang, K, Sherman, W, Heroux, A, Sastry, G.M, Baghbanzadeh, M, Lockett, M.R, Whitesides, G.M. | | Deposit date: | 2015-02-26 | | Release date: | 2015-03-25 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Interactions between Hofmeister Anions and the Binding Pocket of a Protein.
J.Am.Chem.Soc., 137, 2015
|
|
4YGN
 
 | | NaI--Interactions between Hofmeister Anions and the Binding Pocket of a Protein | | Descriptor: | Carbonic anhydrase 2, IODIDE ION, ZINC ION | | Authors: | Fox, J.M, Kang, K, Sherman, W, Heroux, A, Sastry, G.M, Baghbanzadeh, M, Lockett, M.R, Whitesides, G.M. | | Deposit date: | 2015-02-26 | | Release date: | 2015-03-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.23 Å) | | Cite: | Interactions between Hofmeister Anions and the Binding Pocket of a Protein.
J.Am.Chem.Soc., 137, 2015
|
|
4YGL
 
 | | NaClO4--Interactions between Hofmeister Anions and the Binding Pocket of a Protein | | Descriptor: | Carbonic anhydrase 2, HYDROXIDE ION, PERCHLORATE ION, ... | | Authors: | Fox, J.M, Kang, K, Sherman, W, Heroux, A, Sastry, G.M, Baghbanzadeh, M, Lockett, M.R, Whitesides, G.M. | | Deposit date: | 2015-02-26 | | Release date: | 2015-03-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Interactions between Hofmeister Anions and the Binding Pocket of a Protein.
J.Am.Chem.Soc., 137, 2015
|
|
4BGN
 
 | | cryo-EM structure of the NavCt voltage-gated sodium channel | | Descriptor: | VOLTAGE-GATED SODIUM CHANNEL | | Authors: | Tsai, C.J, Tani, K, Irie, K, Hiroaki, Y, Shimomura, T, Mcmillan, D.G, Cook, G.M, Schertler, G, Fujiyoshi, Y, Li, X.D. | | Deposit date: | 2013-03-28 | | Release date: | 2013-07-10 | | Last modified: | 2023-12-20 | | Method: | ELECTRON CRYSTALLOGRAPHY (9 Å) | | Cite: | Two Alternative Conformations of a Voltage-Gated Sodium Channel.
J.Mol.Biol., 425, 2013
|
|
4V8I
 
 | | Crystal structure of YfiA bound to the 70S ribosome. | | Descriptor: | 16S Ribosomal RNA, 23S Ribosomal RNA, 30S Ribosomal Protein S10, ... | | Authors: | Polikanov, Y.S, Blaha, G.M, Steitz, T.A. | | Deposit date: | 2011-12-12 | | Release date: | 2014-07-09 | | Last modified: | 2014-12-10 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | How hibernation factors RMF, HPF, and YfiA turn off protein synthesis.
Science, 336, 2012
|
|
4B7L
 
 | |
4V8H
 
 | | Crystal structure of HPF bound to the 70S ribosome. | | Descriptor: | 16S Ribosomal RNA, 23S Ribosomal RNA, 30S Ribosomal Protein S10, ... | | Authors: | Polikanov, Y.S, Blaha, G.M, Steitz, T.A. | | Deposit date: | 2011-12-11 | | Release date: | 2014-07-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | How Hibernation Factors RMF, HPF, and YfiA Turn Off Protein Synthesis.
Science, 336, 2012
|
|
6I1B
 
 | |
6WTR
 
 | | Structure of Human pir-miRNA-300 Apical Loop Fused to the YdaO Riboswitch Scaffold | | Descriptor: | (2R,3R,3aS,5R,7aR,9R,10R,10aS,12R,14aR)-2,9-bis(6-amino-9H-purin-9-yl)octahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8 ]tetraoxadiphosphacyclododecine-3,5,10,12-tetrol 5,12-dioxide, MAGNESIUM ION, POTASSIUM ION, ... | | Authors: | Shoffner, G.M, Peng, Z, Guo, F. | | Deposit date: | 2020-05-03 | | Release date: | 2021-05-05 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.082 Å) | | Cite: | Structures of microRNA-precursor apical junctions and loops revealed by scaffold-directed crystallography: importance of non-canonical base pairs in processing
To Be Published
|
|
6WTL
 
 | | Structure of Human pir-miRNA-19b-2 Apical Loop and One-base-pair Fused to the YdaO Riboswitch Scaffold | | Descriptor: | (2R,3R,3aS,5R,7aR,9R,10R,10aS,12R,14aR)-2,9-bis(6-amino-9H-purin-9-yl)octahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8 ]tetraoxadiphosphacyclododecine-3,5,10,12-tetrol 5,12-dioxide, MAGNESIUM ION, POTASSIUM ION, ... | | Authors: | Shoffner, G.M, Peng, Z, Guo, F. | | Deposit date: | 2020-05-03 | | Release date: | 2021-05-05 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Three-dimensional structures of pri-miRNA apical junctions and loops revealed by scaffold-directed crystallography
To Be Published
|
|
4UQ3
 
 | | Crystal structure of HLA-A0201 in complex with an azobenzene- containing peptide | | Descriptor: | AZOBENZENE-CONTAINING PEPTIDE, BETA-2-MICROGLOBULIN, HLA CLASS I HISTOCOMPATIBILITY ANTIGEN, ... | | Authors: | Thong, S.Y, Yap, J.W, Lim, P.Y, Verhelst, S.H, Lescar, J, Meijers, R, Grotenbreg, G.M. | | Deposit date: | 2014-06-19 | | Release date: | 2014-09-17 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Bioorthogonal cleavage and exchange of major histocompatibility complex ligands by employing azobenzene-containing peptides.
Angew. Chem. Int. Ed. Engl., 53, 2014
|
|
4UQ2
 
 | | Crystal structure of HLA-A1101 in complex with an azobenzene- containing peptide | | Descriptor: | AZOBENZENE-CONTAINING PEPTIDE, BETA-2-MICROGLOBULIN, HLA CLASS I HISTOCOMPATIBILITY ANTIGEN, ... | | Authors: | Thong, S.Y, Yap, J.W, Lim, P.Y, Verhelst, S.H, Lescar, J, Meijers, R, Grotenbreg, G.M. | | Deposit date: | 2014-06-19 | | Release date: | 2014-09-17 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Bioorthogonal Cleavage and Exchange of Major Histocompatibility Complex Ligands by Employing Azobenzene-Containing Peptides.
Angew.Chem.Int.Ed.Engl., 53, 2014
|
|
4V8G
 
 | | Crystal structure of RMF bound to the 70S ribosome. | | Descriptor: | 16S Ribosomal RNA, 23S Ribosomal RNA, 30S Ribosomal Protein S10, ... | | Authors: | Polikanov, Y.S, Blaha, G.M, Steitz, T.A. | | Deposit date: | 2011-12-11 | | Release date: | 2014-07-09 | | Last modified: | 2014-12-10 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | How hibernation factors RMF, HPF, and YfiA turn off protein synthesis.
Science, 336, 2012
|
|
1EMH
 
 | | CRYSTAL STRUCTURE OF HUMAN URACIL-DNA GLYCOSYLASE BOUND TO UNCLEAVED SUBSTRATE-CONTAINING DNA | | Descriptor: | DNA (5'-D(*AP*AP*AP*GP*AP*TP*AP*AP*CP*A)-3'), DNA (5'-D(*TP*GP*TP*(P2U)P*AP*TP*CP*TP*T)-3'), URACIL-DNA GLYCOSYLASE | | Authors: | Parikh, S.S, Slupphaug, G, Krokan, H.E, Blackburn, G.M, Tainer, J.A. | | Deposit date: | 2000-03-16 | | Release date: | 2000-05-16 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Uracil-DNA glycosylase-DNA substrate and product structures: conformational strain promotes catalytic efficiency by coupled stereoelectronic effects.
Proc.Natl.Acad.Sci.USA, 97, 2000
|
|
1EG7
 
 | | THE CRYSTAL STRUCTURE OF FORMYLTETRAHYDROFOLATE SYNTHETASE FROM MOORELLA THERMOACETICA | | Descriptor: | FORMYLTETRAHYDROFOLATE SYNTHETASE, SULFATE ION | | Authors: | Radfar, R, Shin, R, Sheldrick, G.M, Minor, W, Lovell, C.R, Odom, J.D, Dunlap, R.B, Lebioda, L. | | Deposit date: | 2000-02-14 | | Release date: | 2001-02-14 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The crystal structure of N(10)-formyltetrahydrofolate synthetase from Moorella thermoacetica.
Biochemistry, 39, 2000
|
|
1EPS
 
 | | STRUCTURE AND TOPOLOGICAL SYMMETRY OF THE GLYPHOSPHATE 5-ENOL-PYRUVYLSHIKIMATE-3-PHOSPHATE SYNTHASE: A DISTINCTIVE PROTEIN FOLD | | Descriptor: | 5-ENOL-PYRUVYL-3-PHOSPHATE SYNTHASE | | Authors: | Stallings, W.C, Abdel-Meguid, S.S, Lim, L.W, Shieh, H.-S, Dayringer, H.E, Leimgruber, N.K, Stegeman, R.A, Anderson, K.S, Sikorski, J.A, Padgette, S.R, Kishore, G.M. | | Deposit date: | 1991-04-05 | | Release date: | 1993-07-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure and topological symmetry of the glyphosate target 5-enolpyruvylshikimate-3-phosphate synthase: a distinctive protein fold.
Proc.Natl.Acad.Sci.USA, 88, 1991
|
|
1EMJ
 
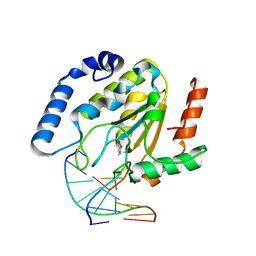 | | URACIL-DNA GLYCOSYLASE BOUND TO DNA CONTAINING A 4'-THIO-2'DEOXYURIDINE ANALOG PRODUCT | | Descriptor: | DNA (5'-D(*AP*AP*AP*GP*AP*TP*AP*AP*CP*A)-3'), DNA (5'-D(*TP*GP*TP*(ASU)P*AP*TP*CP*TP*T)-3'), URACIL, ... | | Authors: | Parikh, S.S, Walcher, G, Jones, G.D, Slupphaug, G, Krokan, H.E, Blackburn, G.M, Tainer, J.A. | | Deposit date: | 2000-03-16 | | Release date: | 2000-05-16 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Uracil-DNA glycosylase-DNA substrate and product structures: conformational strain promotes catalytic efficiency by coupled stereoelectronic effects.
Proc.Natl.Acad.Sci.USA, 97, 2000
|
|
4KH0
 
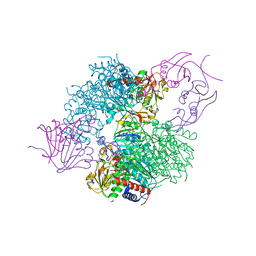 | | The R state structure of E. coli ATCase with ATP and Magnesium bound | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Aspartate carbamoyltransferase, Aspartate carbamoyltransferase regulatory chain, ... | | Authors: | Cockrell, G.M, Zheng, Y, Guo, W, Peterson, A.W, Kantrowitz, E.R. | | Deposit date: | 2013-04-29 | | Release date: | 2013-11-27 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | New Paradigm for Allosteric Regulation of Escherichia coli Aspartate Transcarbamoylase.
Biochemistry, 52, 2013
|
|
4KH1
 
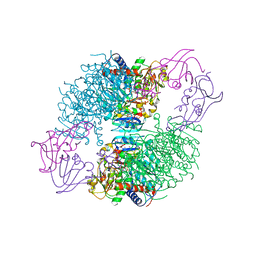 | | The R state structure of E. coli ATCase with CTP,UTP, and Magnesium bound | | Descriptor: | Aspartate carbamoyltransferase, Aspartate carbamoyltransferase regulatory chain, CYTIDINE-5'-TRIPHOSPHATE, ... | | Authors: | Cockrell, G.M, Zheng, Y, Guo, W, Peterson, A.W, Kantrowitz, E.R. | | Deposit date: | 2013-04-29 | | Release date: | 2013-11-27 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | New Paradigm for Allosteric Regulation of Escherichia coli Aspartate Transcarbamoylase.
Biochemistry, 52, 2013
|
|
4KGZ
 
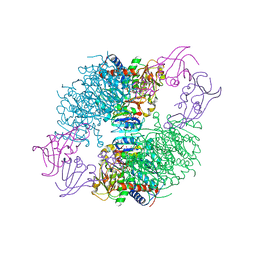 | | The R state structure of E. coli ATCase with UTP and Magnesium bound | | Descriptor: | Aspartate carbamoyltransferase, Aspartate carbamoyltransferase regulatory chain, MAGNESIUM ION, ... | | Authors: | Cockrell, G.M, Zheng, Y, Guo, W, Peterson, A.W, Kantrowitz, E.R. | | Deposit date: | 2013-04-29 | | Release date: | 2013-11-27 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | New Paradigm for Allosteric Regulation of Escherichia coli Aspartate Transcarbamoylase.
Biochemistry, 52, 2013
|
|
3MBU
 
 | | Structure of a bipyridine-modified PNA duplex | | Descriptor: | 1,2-ETHANEDIOL, Bipyridine-PNA, CARBONATE ION, ... | | Authors: | Yeh, J.I, Pohl, E, Truan, D, He, W, Sheldrick, G.M, Du, S, Achim, C. | | Deposit date: | 2010-03-26 | | Release date: | 2011-03-30 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | The crystal structure of non-modified and bipyridine-modified PNA duplexes.
Chemistry, 16, 2010
|
|
