7KR0
 
 | | Crystal structure of SARS-CoV-2 NSP3 macrodomain (C2 crystal form, 100 K) | | Descriptor: | Non-structural protein 3 | | Authors: | Correy, G.J, Young, I.D, Thompson, M.C, Fraser, J.S. | | Deposit date: | 2020-11-18 | | Release date: | 2020-12-09 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (0.77 Å) | | Cite: | Fragment binding to the Nsp3 macrodomain of SARS-CoV-2 identified through crystallographic screening and computational docking.
Sci Adv, 7, 2021
|
|
7L0D
 
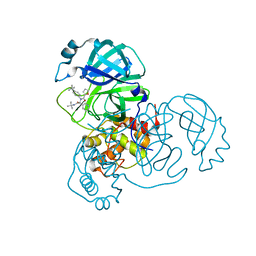 | | SARS-CoV-2 Main Protease (Mpro) in Complex with ML188 | | Descriptor: | 3C-like proteinase, N-[(1R)-2-(tert-butylamino)-2-oxo-1-(pyridin-3-yl)ethyl]-N-(4-tert-butylphenyl)furan-2-carboxamide | | Authors: | Lockbaum, G.J, Lee, J.M, Reyes, A.C, Nalivaika, E.A, Ali, A, Yilmaz, N.K, Thompson, P.R, Schiffer, C.A. | | Deposit date: | 2020-12-11 | | Release date: | 2021-02-10 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Crystal Structure of SARS-CoV-2 Main Protease in Complex with the Non-Covalent Inhibitor ML188.
Viruses, 13, 2021
|
|
6Q2R
 
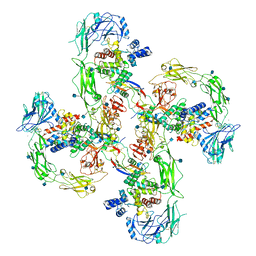 | | Cryo-EM structure of RET/GFRa2/NRTN extracellular complex in the tetrameric form | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, GDNF family receptor alpha-2, ... | | Authors: | Li, J, Shang, G.J, Chen, Y.J, Brautigam, C.A, Liou, J, Zhang, X.W, Bai, X.C. | | Deposit date: | 2019-08-08 | | Release date: | 2019-10-02 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Cryo-EM analyses reveal the common mechanism and diversification in the activation of RET by different ligands.
Elife, 8, 2019
|
|
6PXV
 
 | | Cryo-EM structure of full-length insulin receptor bound to 4 insulin. 3D refinement was focused on the extracellular region. | | Descriptor: | Insulin, Insulin receptor | | Authors: | Uchikawa, E, Choi, E, Shang, G.J, Yu, H.T, Bai, X.C. | | Deposit date: | 2019-07-27 | | Release date: | 2019-09-04 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Activation mechanism of the insulin receptor revealed by cryo-EM structure of the fully liganded receptor-ligand complex.
Elife, 8, 2019
|
|
6Q2J
 
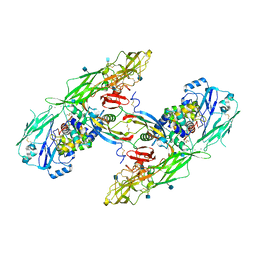 | | Cryo-EM structure of extracellular dimeric complex of RET/GFRAL/GDF15 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, GDNF family receptor alpha-like, ... | | Authors: | Li, J, Shang, G.J, Chen, Y.J, Brautigam, C.A, Liou, J, Zhang, X.W, Bai, X.C. | | Deposit date: | 2019-08-08 | | Release date: | 2019-10-02 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Cryo-EM analyses reveal the common mechanism and diversification in the activation of RET by different ligands.
Elife, 8, 2019
|
|
6Q2S
 
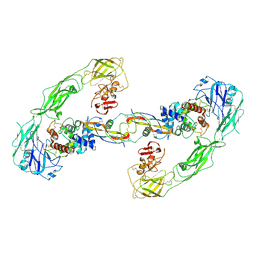 | | Cryo-EM structure of RET/GFRa3/ARTN extracellular complex. The 3D refinement was applied with C2 symmetry. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, GDNF family receptor alpha-3, ... | | Authors: | Li, J, Shang, G.J, Chen, Y.J, Brautigam, C.A, Liou, J, Zhang, X.W, Bai, X.C. | | Deposit date: | 2019-08-08 | | Release date: | 2019-10-02 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Cryo-EM analyses reveal the common mechanism and diversification in the activation of RET by different ligands.
Elife, 8, 2019
|
|
6Q2O
 
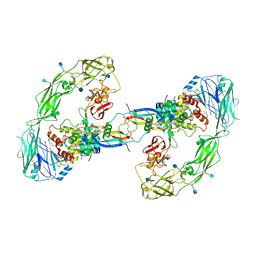 | | Cryo-EM structure of RET/GFRa2/NRTN extracellular complex. The 3D refinement was applied with C2 symmetry. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, GDNF family receptor alpha-2, ... | | Authors: | Li, J, Shang, G.J, Chen, Y.J, Brautigam, C.A, Liou, J, Zhang, X.W, Bai, X.C. | | Deposit date: | 2019-08-08 | | Release date: | 2019-10-02 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.65 Å) | | Cite: | Cryo-EM analyses reveal the common mechanism and diversification in the activation of RET by different ligands.
Elife, 8, 2019
|
|
6PXW
 
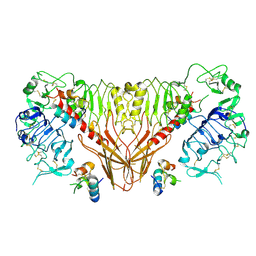 | | Cryo-EM structure of full-length insulin receptor bound to 4 insulin. 3D refinement was focused on the top part of the receptor complex. | | Descriptor: | Insulin, Insulin receptor | | Authors: | Uchikawa, E, Choi, E, Shang, G.J, Yu, H.T, Bai, X.C. | | Deposit date: | 2019-07-28 | | Release date: | 2019-09-04 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Activation mechanism of the insulin receptor revealed by cryo-EM structure of the fully liganded receptor-ligand complex.
Elife, 8, 2019
|
|
6Q2N
 
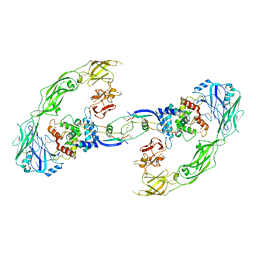 | | Cryo-EM structure of RET/GFRa1/GDNF extracellular complex | | Descriptor: | CALCIUM ION, GDNF family receptor alpha-1, Glial cell line-derived neurotrophic factor, ... | | Authors: | Li, J, Shang, G.J, Chen, Y.J, Brautigam, C.A, Liou, J, Zhang, X.W, Bai, X.C. | | Deposit date: | 2019-08-08 | | Release date: | 2019-10-02 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Cryo-EM analyses reveal the common mechanism and diversification in the activation of RET by different ligands.
Elife, 8, 2019
|
|
7LUP
 
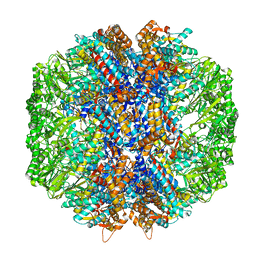 | | Human TRiC/CCT complex with reovirus outer capsid protein sigma-3 | | Descriptor: | Outer capsid protein sigma-3, T-complex protein 1 subunit alpha, T-complex protein 1 subunit beta, ... | | Authors: | Knowlton, J.J, Gestaut, D, Ma, B, Taylor, G, Seven, A.B, Leitner, A, Wilson, G.J, Shanker, S, Yates, N.A, Prasad, B.V.V, Aebersold, R, Chiu, W, Frydman, J, Dermody, T.S. | | Deposit date: | 2021-02-22 | | Release date: | 2021-03-03 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (6.2 Å) | | Cite: | Structural and functional dissection of reovirus capsid folding and assembly by the prefoldin-TRiC/CCT chaperone network.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7LUM
 
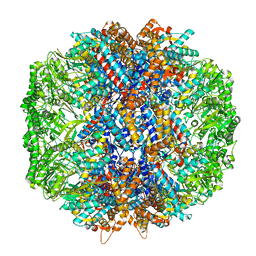 | | Human TRiC in ATP/AlFx closed state | | Descriptor: | T-complex protein 1 subunit alpha, T-complex protein 1 subunit beta, T-complex protein 1 subunit delta, ... | | Authors: | Knowlton, J.J, Gestaut, D, Ma, B, Taylor, G, Seven, A.B, Leitner, A, Wilson, G.J, Shanker, S, Yates, N.A, Prasad, B.V.V, Aebersold, R, Chiu, W, Frydman, J, Dermody, T.S. | | Deposit date: | 2021-02-22 | | Release date: | 2021-03-03 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Structural and functional dissection of reovirus capsid folding and assembly by the prefoldin-TRiC/CCT chaperone network.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7KQO
 
 | |
7LRU
 
 | | Crystal structure of SFPQ-NONO-SFPQ chimeric protein homodimer | | Descriptor: | Splicing factor, proline- and glutamine-rich,Isoform 2 of Non-POU domain-containing octamer-binding protein,Isoform Short of Splicing factor, proline- and glutamine-rich | | Authors: | Marshall, A.C, Bond, C.S, Mohnen, I, Knott, G.J. | | Deposit date: | 2021-02-17 | | Release date: | 2021-05-12 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of SFPQ-NONO-SFPQ chimeric protein homodimer
To Be Published
|
|
7L8I
 
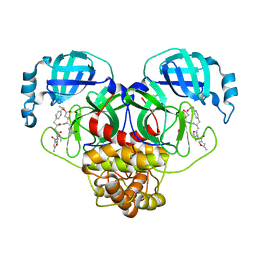 | | SARS-CoV-2 Main Protease (Mpro) in Complex with Rupintrivir (P21) | | Descriptor: | 3C-like proteinase, 4-{2-(4-FLUORO-BENZYL)-6-METHYL-5-[(5-METHYL-ISOXAZOLE-3-CARBONYL)-AMINO]-4-OXO-HEPTANOYLAMINO}-5-(2-OXO-PYRROLIDIN-3-YL)-PENTANOIC ACID ETHYL ESTER | | Authors: | Lockbaum, G.J, Henes, M, Lee, J.M, Timm, J, Nalivaika, E.A, Yilmaz, N.K, Thompson, P.R, Schiffer, C.A. | | Deposit date: | 2020-12-31 | | Release date: | 2021-09-22 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Pan-3C Protease Inhibitor Rupintrivir Binds SARS-CoV-2 Main Protease in a Unique Binding Mode.
Biochemistry, 60, 2021
|
|
7L8J
 
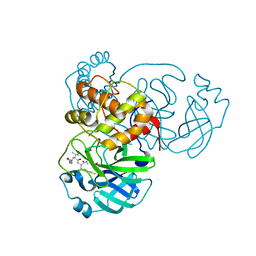 | | SARS-CoV-2 Main Protease (Mpro) in Complex with Rupintrivir (P21212) | | Descriptor: | 3C-like proteinase, 4-{2-(4-FLUORO-BENZYL)-6-METHYL-5-[(5-METHYL-ISOXAZOLE-3-CARBONYL)-AMINO]-4-OXO-HEPTANOYLAMINO}-5-(2-OXO-PYRROLIDIN-3-YL)-PENTANOIC ACID ETHYL ESTER | | Authors: | Lockbaum, G.J, Henes, M, Lee, J.M, Timm, J, Nalivaika, E.A, Yilmaz, N.K, Thompson, P.R, Schiffer, C.A. | | Deposit date: | 2020-12-31 | | Release date: | 2021-09-22 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Pan-3C Protease Inhibitor Rupintrivir Binds SARS-CoV-2 Main Protease in a Unique Binding Mode.
Biochemistry, 60, 2021
|
|
7L8H
 
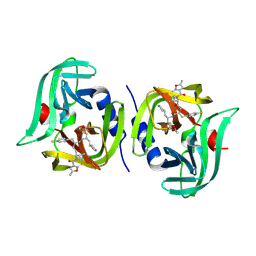 | | EV68 3C protease (3Cpro) in Complex with Rupintrivir | | Descriptor: | 3C Protease, 4-{2-(4-FLUORO-BENZYL)-6-METHYL-5-[(5-METHYL-ISOXAZOLE-3-CARBONYL)-AMINO]-4-OXO-HEPTANOYLAMINO}-5-(2-OXO-PYRROLIDIN-3-YL)-PENTANOIC ACID ETHYL ESTER | | Authors: | Lockbaum, G.J, Henes, M, Lee, J.M, Timm, J, Nalivaika, E.A, Yilmaz, N.K, Thompson, P.R, Schiffer, C.A. | | Deposit date: | 2020-12-31 | | Release date: | 2021-09-22 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Pan-3C Protease Inhibitor Rupintrivir Binds SARS-CoV-2 Main Protease in a Unique Binding Mode.
Biochemistry, 60, 2021
|
|
2WCO
 
 | | Structures of the Streptomyces coelicolor A3(2) Hyaluronan Lyase in Complex with Oligosaccharide Substrates and an Inhibitor | | Descriptor: | 4-deoxy-beta-D-glucopyranuronic acid-(1-3)-2-acetamido-2-deoxy-beta-D-glucopyranose, FORMIC ACID, GLYCEROL, ... | | Authors: | Elmabrouk, Z.H, Taylor, E.J, Vincent, F, Smith, N.L, Zhang, M, Charnock, S.J, Turkenburg, J.P, Davies, G.J, Black, G.W. | | Deposit date: | 2009-03-12 | | Release date: | 2010-08-18 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Crystal Structures of a Family 8 Polysaccharide Lyase Reveal Open and Highly Occluded Substrate-Binding Cleft Conformations.
Proteins, 79, 2011
|
|
2WDA
 
 | | The X-ray structure of the Streptomyces coelicolor A3 Chondroitin AC Lyase in Complex with Chondroitin sulphate | | Descriptor: | 4-deoxy-alpha-L-threo-hex-4-enopyranuronic acid-(1-3)-2-acetamido-2-deoxy-4-O-sulfo-beta-D-galactopyranose, DI(HYDROXYETHYL)ETHER, FORMIC ACID, ... | | Authors: | Elmabrouk, Z.H, Taylor, E.J, Vincent, F, Smith, N.L, Turkenburg, J.P, Davies, G.J, Black, G.W. | | Deposit date: | 2009-03-23 | | Release date: | 2010-04-21 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structures of a Family 8 Polysaccharide Lyase Reveal Open and Highly Occluded Substrate-Binding Cleft Conformations.
Proteins, 79, 2011
|
|
2IV9
 
 | |
2IV8
 
 | |
2JI8
 
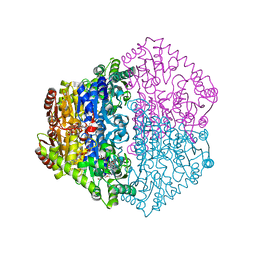 | | X-ray structure of Oxalyl-CoA decarboxylase in complex with Formyl- CoA | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, OXALYL-COA DECARBOXYLASE, ... | | Authors: | Berthold, C.L, Toyota, C.G, Moussatche, P, Wood, M.D, Leeper, F, Richards, N.G.J, Lindqvist, Y. | | Deposit date: | 2007-02-26 | | Release date: | 2007-07-17 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystallographic Snapshots of Oxalyl-Coa Decarboxylase Give Insights Into Catalysis by Nonoxidative Thdp-Dependent Decarboxylases
Structure, 15, 2007
|
|
2WQ8
 
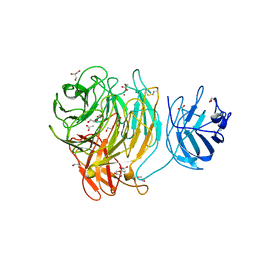 | | Glycan labelling using engineered variants of galactose oxidase obtained by directed evolution | | Descriptor: | 1,2-ETHANEDIOL, ACETIC ACID, CALCIUM ION, ... | | Authors: | Rannes, J.G, Ioannou, A, Willies, S.C, Behrens, C, Grogan, G.J, Flitsch, S.L, Turner, N.J. | | Deposit date: | 2009-08-14 | | Release date: | 2010-09-01 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Glycoprotein Labeling Using Engineered Variants of Galactose Oxidase Obtained by Directed Evolution.
J.Am.Chem.Soc., 133, 2011
|
|
2JI7
 
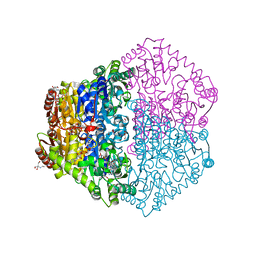 | | X-ray structure of Oxalyl-CoA decarboxylase with covalent reaction intermediate | | Descriptor: | 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 3-[(4-AMINO-2-METHYLPYRIMIDIN-5-YL)METHYL]-2-{(1R,11R,15S,17R)-19-[(2R,3S,4R,5R)-5-(6-AMINO-9H-PURIN-9-YL)-4-HYDROXY-3-(PHOSPHONOOXY)TETRAHYDROFURAN-2-YL]-1,11,15,17-TETRAHYDROXY-12,12-DIMETHYL-15,17-DIOXIDO-6,10-DIOXO-14,16,18-TRIOXA-2-THIA-5,9-DIAZA-15,17-DIPHOSPHANONADEC-1-YL}-5-(2-{[(R)-HYDROXY(PHOSPHONOOXY)PHOSPHORYL]OXY}ETHYL)-4-METHYL-1,3-THIAZOL-3-IUM, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Berthold, C.L, Toyota, C.G, Moussatche, P, Wood, M.D, Leeper, F, Richards, N.G.J, Lindqvist, Y. | | Deposit date: | 2007-02-26 | | Release date: | 2007-07-17 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Crystallographic Snapshots of Oxalyl-Coa Decarboxylase Give Insights Into Catalysis by Nonoxidative Thdp-Dependent Decarboxylases
Structure, 15, 2007
|
|
2X03
 
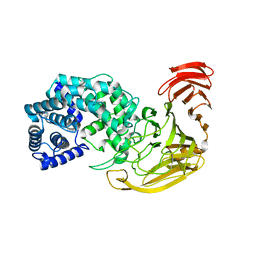 | | The X-ray structure of the Streptomyces coelicolor A3 Chondroitin AC Lyase Y253A mutant | | Descriptor: | MAGNESIUM ION, PUTATIVE SECRETED LYASE | | Authors: | Elmabrouk, Z.H, Taylor, E.J, Vincent, F, Smith, N.L, Turkenburg, J.P, Davies, G.J, Black, G.W. | | Deposit date: | 2009-12-04 | | Release date: | 2010-08-18 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structures of a Family 8 Polysaccharide Lyase Reveal Open and Highly Occluded Substrate-Binding Cleft Conformations.
Proteins, 79, 2011
|
|
2HUO
 
 | | Crystal structure of mouse myo-inositol oxygenase in complex with substrate | | Descriptor: | 1,2,3,4,5,6-HEXAHYDROXY-CYCLOHEXANE, FE (III) ION, FORMIC ACID, ... | | Authors: | Brown, P.M, Caradoc-Davies, T.T, Dickson, J.M.J, Cooper, G.J.S, Loomes, K.M, Baker, E.N. | | Deposit date: | 2006-07-27 | | Release date: | 2006-09-26 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of a substrate complex of myo-inositol oxygenase, a di-iron oxygenase with a key role in inositol metabolism.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
