3MP3
 
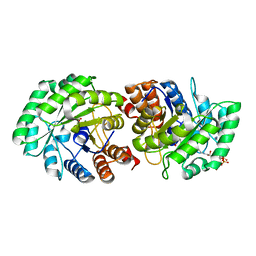 | | Crystal Structure of Human Lyase in complex with inhibitor HG-CoA | | Descriptor: | (3R,5S,9R,21S)-1-[(2R,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-4-hydroxy-3-(phosphonooxy)tetrahydrofuran-2-yl]-3,5,9,21-tetrahydroxy-8,8-dimethyl-10,14,19-trioxo-2,4,6-trioxa-18-thia-11,15-diaza-3,5-diphosphatricosan-23-oic acid 3,5-dioxide, 3-HYDROXYPENTANEDIOIC ACID, Hydroxymethylglutaryl-CoA lyase, ... | | Authors: | Fu, Z, Runquist, J.A, Montgomery, C, Miziorko, H.M, Kim, J.-J.P. | | Deposit date: | 2010-04-24 | | Release date: | 2010-06-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Functional insights into human HMG-CoA lyase from structures of Acyl-CoA-containing ternary complexes.
J.Biol.Chem., 285, 2010
|
|
3N7L
 
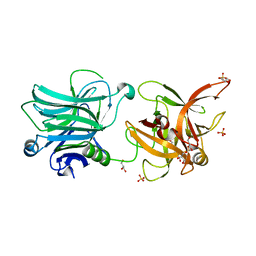 | | Crystal structure of botulinum neurotoxin serotype D/C VPI 5993 binding domain | | Descriptor: | GLYCEROL, Neurotoxin, SULFATE ION | | Authors: | Fu, Z, Karalewitz, A, Kroken, A, Baldwin, M.R, Barbieri, J.T, Kim, J.-J.P. | | Deposit date: | 2010-05-27 | | Release date: | 2010-09-08 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Identification of a Unique Ganglioside Binding Loop within Botulinum Neurotoxins C and D-SA .
Biochemistry, 49, 2010
|
|
1A1X
 
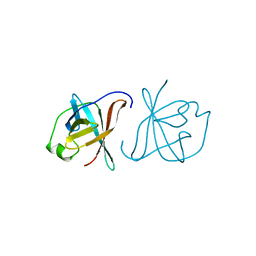 | | CRYSTAL STRUCTURE OF MTCP-1 INVOLVED IN T CELL MALIGNANCIES | | Descriptor: | HMTCP-1 | | Authors: | Fu, Z.Q, Dubois, G.C, Song, S.P, Kulikovskaya, I, Virgilio, L, Rothstein, J, Croce, C.M, Weber, I.T, Harrison, R.W. | | Deposit date: | 1997-12-18 | | Release date: | 1998-05-27 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of MTCP-1: implications for role of TCL-1 and MTCP-1 in T cell malignancies.
Proc.Natl.Acad.Sci.USA, 95, 1998
|
|
7ZOX
 
 | | Nup93 in complex with xhNup93-Nb4i and xNup93-Nb2t | | Descriptor: | Nuclear pore complex protein Nup93, xNup93-Nb2t, xhNup93-Nb4i | | Authors: | Fu, Z, Guttler, T, Colom, M.S. | | Deposit date: | 2022-04-26 | | Release date: | 2023-11-08 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | A checkpoint function for Nup98 in nuclear pore formation suggested by novel inhibitory nanobodies.
Embo J., 2024
|
|
1XVA
 
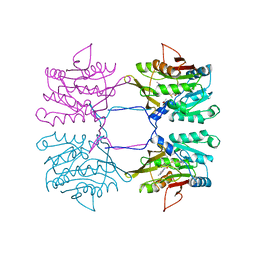 | | METHYLTRANSFERASE | | Descriptor: | ACETATE ION, GLYCINE N-METHYLTRANSFERASE, S-ADENOSYLMETHIONINE | | Authors: | Fu, Z, Hu, Y, Konishi, K, Takata, Y, Ogawa, H, Gomi, T, Fujioka, M, Takusagawa, F. | | Deposit date: | 1996-07-20 | | Release date: | 1997-01-27 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of glycine N-methyltransferase from rat liver.
Biochemistry, 35, 1996
|
|
2P5U
 
 | | Crystal structure of Thermus thermophilus HB8 UDP-glucose 4-epimerase complex with NAD | | Descriptor: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, UDP-glucose 4-epimerase | | Authors: | Fu, Z.-Q, Chen, L, Ebihara, A, Shinkai, A, Kuramitsu, S, Yokoyama, S, Zhao, M, Dillard, B, Chrzas, J, Rose, J.P, Wang, B.-C, Southeast Collaboratory for Structural Genomics (SECSG), RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-03-16 | | Release date: | 2007-04-17 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Crystal structure of Thermus thermophilus HB8 UDP-glucose 4-epimerase complex with NAD
To be Published
|
|
2P5Y
 
 | | Crystal structure of Thermus thermophilus HB8 UDP-glucose 4-epimerase complex with NAD | | Descriptor: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, UDP-glucose 4-epimerase | | Authors: | Fu, Z.-Q, Chen, L, Ebihara, A, Shinkai, A, Kuramitsu, S, Yokoyama, S, Zhu, J, Swindell, J.T, Chrzas, J, Rose, J.P, Wang, B.-C, Southeast Collaboratory for Structural Genomics (SECSG), RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-03-16 | | Release date: | 2007-04-17 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Crystal structure of Thermus thermophilus HB8 UDP-glucose 4-epimerase complex with NAD
To be Published
|
|
2P62
 
 | | Crystal structure of hypothetical protein PH0156 from Pyrococcus horikoshii OT3 | | Descriptor: | Hypothetical protein PH0156 | | Authors: | Fu, Z.-Q, Chen, L, Zhu, J, Swindell, J.T, Ebihara, A, Shinkai, A, Kuramitsu, S, Yokoyama, S, Chrzas, J, Rose, J.P, Wang, B.-C, Southeast Collaboratory for Structural Genomics (SECSG), RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-03-16 | | Release date: | 2007-04-17 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of hypothetical protein PH0156 from Pyrococcus horikoshii OT3
To be Published
|
|
4IQP
 
 | | Crystal Structure of HCRA-W1266A | | Descriptor: | Botulinum neurotoxin type A, GLYCEROL | | Authors: | Fu, Z, Kroken, A.R, Barbieri, J.T, Kim, J.-J.P. | | Deposit date: | 2013-01-12 | | Release date: | 2013-05-29 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Enhancing the Protective Immune Response against Botulism.
Infect.Immun., 81, 2013
|
|
6OT3
 
 | | RF2 accommodated state bound Release complex 70S at 24 ms | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Fu, Z, Indrisiunaite, G, Kaledhonkar, S, Shah, B, Sun, M, Chen, B, Grassucci, R.A, Ehrenberg, M, Frank, J. | | Deposit date: | 2019-05-02 | | Release date: | 2019-06-19 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | The structural basis for release-factor activation during translation termination revealed by time-resolved cryogenic electron microscopy.
Nat Commun, 10, 2019
|
|
6ORL
 
 | | RF1 pre-accommodated 70S complex at 24 ms | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Fu, Z, Indrisiunaite, G, Kaledhonkar, S, Shah, B, Sun, M, Chen, B, Grassucci, R.A, Ehrenberg, M, Frank, J. | | Deposit date: | 2019-04-30 | | Release date: | 2019-06-19 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | The structural basis for release-factor activation during translation termination revealed by time-resolved cryogenic electron microscopy.
Nat Commun, 10, 2019
|
|
6OST
 
 | | RF2 pre-accommodated state bound Release complex 70S at 24ms | | Descriptor: | 16S Ribosomal RNA, 23S Ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Fu, Z, Indrisiunaite, G, Kaledhonkar, S, Shah, B, Sun, M, Chen, B, Grassucci, R.A, Ehrenberg, M, Frank, J. | | Deposit date: | 2019-05-02 | | Release date: | 2019-06-19 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | The structural basis for release-factor activation during translation termination revealed by time-resolved cryogenic electron microscopy.
Nat Commun, 10, 2019
|
|
1ZTD
 
 | | Hypothetical Protein Pfu-631545-001 From Pyrococcus furiosus | | Descriptor: | Hypothetical Protein Pfu-631545-001 | | Authors: | Fu, Z.-Q, Horanyi, P, Florence, Q, Liu, Z.-J, Chen, L, Lee, D, Habel, J, Xu, H, Nguyen, D, Chang, S.-H, Zhou, W, Zhang, H, Jenney Jr, F.E, Sha, B, Adams, M.W.W, Rose, J.P, Wang, B.-C, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2005-05-26 | | Release date: | 2005-06-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Hypothetical Protein Pfu-631545-001 From Pyrococcus furiosus
To be Published
|
|
7YW1
 
 | | crystal structure of UBE2O | | Descriptor: | (E3-independent) E2 ubiquitin-conjugating enzyme UBE2O | | Authors: | Fu, Z, Zhu, W, Huang, H. | | Deposit date: | 2022-08-20 | | Release date: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.27 Å) | | Cite: | crystal structure of UBE2O
To Be Published
|
|
2FZF
 
 | | Hypothetical Protein Pfu-1136390-001 From Pyrococcus furiosus | | Descriptor: | hypothetical protein | | Authors: | Fu, Z.-Q, Liu, Z.-J, Lee, D, Kelley, L, Chen, L, Tempel, W, Shah, N, Horanyi, P, Lee, H.S, Habel, J, Dillard, B.D, Nguyen, D, Chang, S.-H, Zhang, H, Chang, J, Sugar, F.J, Poole, F.L, Jenney Jr, F.E, Adams, M.W.W, Rose, J.P, Wang, B.-C, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2006-02-09 | | Release date: | 2006-02-21 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Hypothetical Protein Pfu-1136390-001 From Pyrococcus furiosus
To be published
|
|
6DV2
 
 | | Crystal Structure of Human Mitochondrial Trifunctional Protein | | Descriptor: | Trifunctional enzyme subunit alpha, mitochondrial, Trifunctional enzyme subunit beta | | Authors: | Fu, Z, Xia, C, Battaile, K.P, Kim, J.P. | | Deposit date: | 2018-06-22 | | Release date: | 2018-09-26 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Crystal structure of human mitochondrial trifunctional protein, a fatty acid beta-oxidation metabolon.
Proc. Natl. Acad. Sci. U.S.A., 116, 2019
|
|
5YL4
 
 | | CRYSTAL STRUCTURE OF T2R-TTL-8WR COMPLEX | | Descriptor: | (3Z,6Z)-3-[(4-tert-butyl-1H-imidazol-5-yl)methylidene]-6-[[3-(phenylcarbonyl)phenyl]methylidene]piperazine-2,5-dione, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, ... | | Authors: | Fu, Z.Y, Li, W.B, Chu, Y.Y, Hou, Y.W, Ji, C.P. | | Deposit date: | 2017-10-17 | | Release date: | 2017-11-15 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.64 Å) | | Cite: | Design, synthesis and biological activity evaluation of plinabulin derivatives based on co-crystal structure
To Be Published
|
|
7DG6
 
 | |
8GXR
 
 | |
7CJM
 
 | | SARS CoV-2 PLpro in complex with GRL0617 | | Descriptor: | 5-amino-2-methyl-N-[(1R)-1-naphthalen-1-ylethyl]benzamide, Non-structural protein 3, ZINC ION | | Authors: | Fu, Z, Huang, H. | | Deposit date: | 2020-07-11 | | Release date: | 2020-09-02 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | The complex structure of GRL0617 and SARS-CoV-2 PLpro reveals a hot spot for antiviral drug discovery.
Nat Commun, 12, 2021
|
|
2PG0
 
 | | Crystal structure of acyl-CoA dehydrogenase from Geobacillus kaustophilus | | Descriptor: | Acyl-CoA dehydrogenase, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Chen, L, Chen, L.-Q, Ebihara, A, Shinkai, A, Kuramitsu, S, Yokoyama, S, Zhao, M, Li, Y, Fu, Z.-Q, Rose, J.P, Wang, B.-C, Southeast Collaboratory for Structural Genomics (SECSG), RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-04-06 | | Release date: | 2007-05-08 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of acyl-CoA dehydrogenase from G. kaustophilus
To be Published
|
|
1MLD
 
 | | REFINED STRUCTURE OF MITOCHONDRIAL MALATE DEHYDROGENASE FROM PORCINE HEART AND THE CONSENSUS STRUCTURE FOR DICARBOXYLIC ACID OXIDOREDUCTASES | | Descriptor: | CITRIC ACID, MALATE DEHYDROGENASE | | Authors: | Gleason, W.B, Fu, Z, Birktoft, J.J, Banaszak, L.J. | | Deposit date: | 1994-01-24 | | Release date: | 1995-01-26 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Refined crystal structure of mitochondrial malate dehydrogenase from porcine heart and the consensus structure for dicarboxylic acid oxidoreductases.
Biochemistry, 33, 1994
|
|
7KJX
 
 | | Structure of HIV-1 reverse transcriptase initiation complex core with nevirapine | | Descriptor: | 11-CYCLOPROPYL-5,11-DIHYDRO-4-METHYL-6H-DIPYRIDO[3,2-B:2',3'-E][1,4]DIAZEPIN-6-ONE, HIV-1 viral RNA fragment, MAGNESIUM ION, ... | | Authors: | Ha, B, Larsen, K.P, Zhang, J, Fu, Z, Montabana, E, Jackson, L.N, Chen, D.H, Puglisi, E.V. | | Deposit date: | 2020-10-26 | | Release date: | 2021-03-17 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | High-resolution view of HIV-1 reverse transcriptase initiation complexes and inhibition by NNRTI drugs.
Nat Commun, 12, 2021
|
|
7KJV
 
 | | Structure of HIV-1 reverse transcriptase initiation complex core | | Descriptor: | HIV-1 viral RNA fragment, MAGNESIUM ION, Reverse transcriptase/ribonuclease H, ... | | Authors: | Ha, B, Larsen, K.P, Zhang, J, Fu, Z, Montabana, E, Jackson, L.N, Chen, D.H, Puglisi, E.V. | | Deposit date: | 2020-10-26 | | Release date: | 2021-03-17 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | High-resolution view of HIV-1 reverse transcriptase initiation complexes and inhibition by NNRTI drugs.
Nat Commun, 12, 2021
|
|
7KJW
 
 | | Structure of HIV-1 reverse transcriptase initiation complex core with efavirenz | | Descriptor: | (-)-6-CHLORO-4-CYCLOPROPYLETHYNYL-4-TRIFLUOROMETHYL-1,4-DIHYDRO-2H-3,1-BENZOXAZIN-2-ONE, HIV-1 viral RNA fragment, MAGNESIUM ION, ... | | Authors: | Ha, B, Larsen, K.P, Zhang, J, Fu, Z, Montabana, E, Jackson, L.N, Chen, D.H, Puglisi, E.V. | | Deposit date: | 2020-10-26 | | Release date: | 2021-03-17 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | High-resolution view of HIV-1 reverse transcriptase initiation complexes and inhibition by NNRTI drugs.
Nat Commun, 12, 2021
|
|
