2Q2J
 
 | |
1GOD
 
 | | MONOMERIC LYS-49 PHOSPHOLIPASE A2 HOMOLOGUE ISOLATED FROM THE VENOM OF CERROPHIDION (BOTHROPS) GODMANI | | Descriptor: | PROTEIN (PHOSPHOLIPASE A2) | | Authors: | Arni, R.K, Fontes, M.R.M, Barberato, C, Gutierrez, J.M, Diaz-Oreiro, C, Ward, R.J. | | Deposit date: | 1999-04-16 | | Release date: | 1999-04-23 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of myotoxin II, a monomeric Lys49-phospholipase A2 homologue isolated from the venom of Cerrophidion (Bothrops) godmani.
Arch.Biochem.Biophys., 366, 1999
|
|
1HOR
 
 | | STRUCTURE AND CATALYTIC MECHANISM OF GLUCOSAMINE 6-PHOSPHATE DEAMINASE FROM ESCHERICHIA COLI AT 2.1 ANGSTROMS RESOLUTION | | Descriptor: | 2-DEOXY-2-AMINO GLUCITOL-6-PHOSPHATE, GLUCOSAMINE 6-PHOSPHATE DEAMINASE, PHOSPHATE ION | | Authors: | Oliva, G, Fontes, M.R.M, Garratt, R.C, Altamirano, M.M, Calcagno, M.L, Horjales, E. | | Deposit date: | 1995-09-13 | | Release date: | 1996-01-29 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure and catalytic mechanism of glucosamine 6-phosphate deaminase from Escherichia coli at 2.1 A resolution.
Structure, 3, 1995
|
|
4K06
 
 | | Crystal structure of MTX-II from Bothrops brazili venom complexed with polyethylene glycol | | Descriptor: | 2-(2-(2-(2-(2-(2-ETHOXYETHOXY)ETHOXY)ETHOXY)ETHOXY)ETHOXY)ETHANOL, 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, MTX-II, ... | | Authors: | Fernandes, C.A.H, Comparetti, E.J, Borges, R.J, Fontes, M.R.M. | | Deposit date: | 2013-04-03 | | Release date: | 2013-11-13 | | Last modified: | 2013-11-27 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Structural bases for a complete myotoxic mechanism: Crystal structures of two non-catalytic phospholipases A2-like from Bothrops brazili venom.
Biochim.Biophys.Acta, 1834, 2013
|
|
4K09
 
 | |
3HZW
 
 | | Crystal structure of bothropstoxin-I chemically modified by p-bromophenacyl bromide (BPB) | | Descriptor: | ISOPROPYL ALCOHOL, Phospholipase A2 homolog bothropstoxin-1, p-Bromophenacyl bromide | | Authors: | Fernandes, C.A.H, Marchi-Salvador, D.P, Soares, A.M, Fontes, M.R.M. | | Deposit date: | 2009-06-24 | | Release date: | 2010-05-12 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Comparison between apo and complexed structures of bothropstoxin-I reveals the role of Lys122 and Ca(2+)-binding loop region for the catalytically inactive Lys49-PLA(2)s.
J.Struct.Biol., 171, 2010
|
|
3I3I
 
 | |
3I03
 
 | | Crystal structure of bothropstoxin-I chemically modified by p-bromophenacyl bromide (BPB) - monomeric form at a high resolution | | Descriptor: | ISOPROPYL ALCOHOL, Phospholipase A2 homolog bothropstoxin-1, p-Bromophenacyl bromide | | Authors: | Marchi-Salvador, D.P, Fernandes, C.A.H, Soares, A.M, Fontes, M.R.M. | | Deposit date: | 2009-06-24 | | Release date: | 2010-05-12 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Comparison between apo and complexed structures of bothropstoxin-I reveals the role of Lys122 and Ca(2+)-binding loop region for the catalytically inactive Lys49-PLA(2)s.
J.Struct.Biol., 171, 2010
|
|
3HZD
 
 | | Crystal structure of bothropstoxin-I (BthTX-I), a PLA2 homologue from Bothrops jararacussu venom | | Descriptor: | LITHIUM ION, Phospholipase A2 homolog bothropstoxin-1 | | Authors: | Silva, M.C.O, Marchi-Salvador, D.P, Fernandes, C.A.H, Soares, A.M, Fontes, M.R.M. | | Deposit date: | 2009-06-23 | | Release date: | 2009-07-07 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Comparison between apo and complexed structures of bothropstoxin-I reveals the role of Lys122 and Ca(2+)-binding loop region for the catalytically inactive Lys49-PLA(2)s.
J.Struct.Biol., 171, 2010
|
|
5TFV
 
 | |
6B80
 
 | | Crystal structure of myotoxin II from Bothrops moojeni complexed to myristic acid | | Descriptor: | 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, Basic phospholipase A2 homolog 2, MYRISTIC ACID, ... | | Authors: | Salvador, G.H.M, dos Santos, J.I, Fontes, M.R.M. | | Deposit date: | 2017-10-05 | | Release date: | 2018-10-03 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.949 Å) | | Cite: | Structural evidence for a fatty acid-independent myotoxic mechanism for a phospholipase A2-like toxin.
Biochim Biophys Acta Proteins Proteom, 1866, 2018
|
|
6B81
 
 | |
5U5P
 
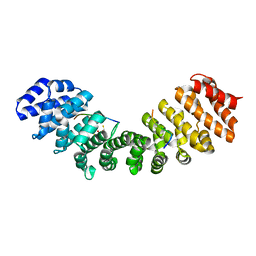 | | Crystal Structure and X-ray Diffraction Data Collection of Importin-alpha from Mus Musculus Complexed with a MLH1 NLS Peptide | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, DNA mismatch repair protein Mlh1, Importin subunit alpha-1 | | Authors: | Barros, A.C, Takeda, A.A, Dreyer, T.R, Velazquez-Campoy, A, Kobe, B, Fontes, M.R. | | Deposit date: | 2016-12-07 | | Release date: | 2018-03-14 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.171 Å) | | Cite: | DNA mismatch repair proteins MLH1 and PMS2 can be imported to the nucleus by a classical nuclear import pathway.
Biochimie, 146, 2018
|
|
6B83
 
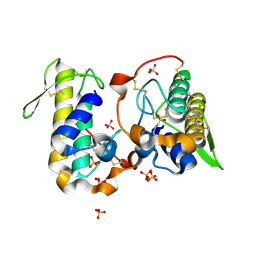 | |
4YU7
 
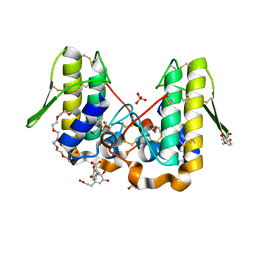 | | Crystal structure of Piratoxin I (PrTX-I) complexed to caffeic acid | | Descriptor: | 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, Basic phospholipase A2 homolog piratoxin-1, CAFFEIC ACID, ... | | Authors: | Fernandes, C.A.H, Fontes, M.R.M. | | Deposit date: | 2015-03-18 | | Release date: | 2015-08-05 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.647 Å) | | Cite: | Structural Basis for the Inhibition of a Phospholipase A2-Like Toxin by Caffeic and Aristolochic Acids.
Plos One, 10, 2015
|
|
5U5R
 
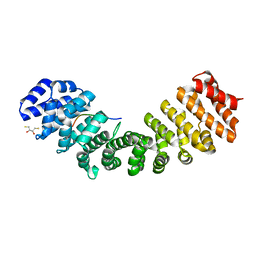 | | Crystal Structure and X-ray Diffraction Data Collection of Importin-alpha from Mus musculus Complexed with a PMS2 NLS Peptide | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, Importin subunit alpha-1, Mismatch repair endonuclease PMS2 | | Authors: | Barros, A.C, Takeda, A.A, Dreyer, T.R, Velazquez-Campoy, A, Kobe, B, Fontes, M.R. | | Deposit date: | 2016-12-07 | | Release date: | 2018-03-14 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | DNA mismatch repair proteins MLH1 and PMS2 can be imported to the nucleus by a classical nuclear import pathway.
Biochimie, 146, 2018
|
|
6CE2
 
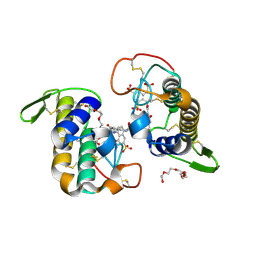 | | Crystal structure of Myotoxin I (MjTX-I) from Bothrops moojeni complexed to inhibitor suramin | | Descriptor: | 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, 8,8'-[CARBONYLBIS[IMINO-3,1-PHENYLENECARBONYLIMINO(4-METHYL-3,1-PHENYLENE)CARBONYLIMINO]]BIS-1,3,5-NAPHTHALENETRISULFON IC ACID, Basic phospholipase A2 homolog 1 | | Authors: | Salvador, G.H.M, Fontes, M.R.M. | | Deposit date: | 2018-02-10 | | Release date: | 2018-07-25 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural and functional characterization of suramin-bound MjTX-I from Bothrops moojeni suggests a particular myotoxic mechanism.
Sci Rep, 8, 2018
|
|
3I3H
 
 | |
5VFN
 
 | |
5VFH
 
 | |
5VFJ
 
 | |
5VQI
 
 | |
5VFM
 
 | |
4YV5
 
 | | Crystal Structure of Myotoxin II from Bothrops moojeni complexed to Suramin | | Descriptor: | 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, 8,8'-[CARBONYLBIS[IMINO-3,1-PHENYLENECARBONYLIMINO(4-METHYL-3,1-PHENYLENE)CARBONYLIMINO]]BIS-1,3,5-NAPHTHALENETRISULFON IC ACID, Basic phospholipase A2 homolog 2, ... | | Authors: | Salvador, G.H.M, Fontes, M.R.M. | | Deposit date: | 2015-03-19 | | Release date: | 2015-10-14 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural and functional evidence for membrane docking and disruption sites on phospholipase A2-like proteins revealed by complexation with the inhibitor suramin.
Acta Crystallogr. D Biol. Crystallogr., 71, 2015
|
|
4YZ7
 
 | | Crystal structure of Piratoxin I (PrTX-I) complexed to aristolochic acid | | Descriptor: | 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, 9-HYDROXY ARISTOLOCHIC ACID, Basic phospholipase A2 homolog piratoxin-1, ... | | Authors: | Fernandes, C.A.H, Fontes, M.R.M. | | Deposit date: | 2015-03-24 | | Release date: | 2015-08-05 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.9589 Å) | | Cite: | Structural Basis for the Inhibition of a Phospholipase A2-Like Toxin by Caffeic and Aristolochic Acids.
Plos One, 10, 2015
|
|
