3CXF
 
 | | Crystal structure of transthyretin variant Y114H | | Descriptor: | Transthyretin | | Authors: | Cendron, L, Zanotti, G, Folli, C, Alfieri, B, Pasquato, N, Berni, R. | | Deposit date: | 2008-04-24 | | Release date: | 2009-04-07 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural and mutational analyses of protein-protein interactions between transthyretin and retinol-binding protein.
Febs J., 275, 2008
|
|
3DO4
 
 | |
2G4G
 
 | |
3DJS
 
 | |
3DJZ
 
 | |
3DK2
 
 | |
5AKT
 
 | | Transthyretin binding heterogeneity and anti-amyloidogenic activity of natural polyphenols and their metabolites: resveratrol-4'-O- glucuronide | | Descriptor: | Resveratrol-4'-O-glucuronide, SULFATE ION, TRANSTHYRETIN | | Authors: | Florio, P, Folli, C, Cianci, M, Del Rio, D, Zanotti, G, Berni, R. | | Deposit date: | 2015-03-05 | | Release date: | 2015-10-21 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Transthyretin Binding Heterogeneity and Anti-Amyloidogenic Activity of Natural Polyphenols and Their Metabolites
J.Biol.Chem., 290, 2015
|
|
3DJT
 
 | |
3DK0
 
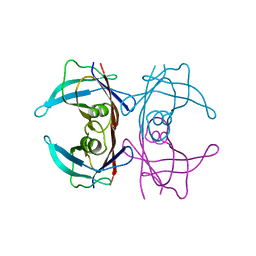 | |
3DJR
 
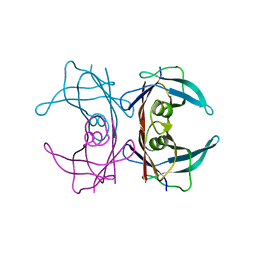 | |
5CR1
 
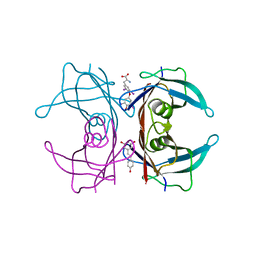 | | Crystal structure of TTR/resveratrol/T4 complex | | Descriptor: | 3,5,3',5'-TETRAIODO-L-THYRONINE, RESVERATROL, Transthyretin | | Authors: | Zanotti, G, Florio, P, Folli, C, Cianci, M, Del Rio, D, Berni, R. | | Deposit date: | 2015-07-22 | | Release date: | 2015-10-21 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.545 Å) | | Cite: | Transthyretin Binding Heterogeneity and Anti-amyloidogenic Activity of Natural Polyphenols and Their Metabolites.
J.Biol.Chem., 290, 2015
|
|
2O73
 
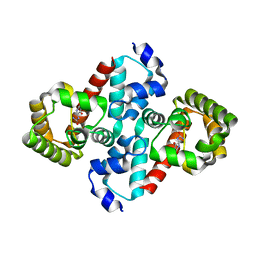 | | Structure of OHCU decarboxylase in complex with allantoin | | Descriptor: | 1-(2,5-DIOXO-2,5-DIHYDRO-1H-IMIDAZOL-4-YL)UREA, OHCU decarboxylase | | Authors: | Cendron, L, Berni, R, Folli, C, Ramazzina, I, Percudani, R, Zanotti, G. | | Deposit date: | 2006-12-10 | | Release date: | 2007-04-10 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The structure of 2-oxo-4-hydroxy-4-carboxy-5-ureidoimidazoline decarboxylase provides insights into the mechanism of uric acid degradation.
J.Biol.Chem., 282, 2007
|
|
2O74
 
 | | Structure of OHCU decarboxylase in complex with guanine | | Descriptor: | GUANINE, OHCU decarboxylase | | Authors: | Cendron, L, Berni, R, Folli, C, Ramazzina, I, Percudani, R, Zanotti, G. | | Deposit date: | 2006-12-10 | | Release date: | 2007-04-10 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The structure of 2-oxo-4-hydroxy-4-carboxy-5-ureidoimidazoline decarboxylase provides insights into the mechanism of uric acid degradation.
J.Biol.Chem., 282, 2007
|
|
2O70
 
 | | Structure of OHCU decarboxylase from zebrafish | | Descriptor: | OHCU decarboxylase | | Authors: | Cendron, L, Berni, R, Folli, C, Ramazzina, I, Percudani, R, Zanotti, G. | | Deposit date: | 2006-12-09 | | Release date: | 2007-04-10 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The structure of 2-oxo-4-hydroxy-4-carboxy-5-ureidoimidazoline decarboxylase provides insights into the mechanism of uric acid degradation.
J.Biol.Chem., 282, 2007
|
|
2NOY
 
 | | Crystal structure of transthyretin mutant I84S at PH 7.5 | | Descriptor: | Transthyretin | | Authors: | Pasquato, N, Berni, R, Folli, C, Alfieri, B, Cendron, L, Zanotti, G. | | Deposit date: | 2006-10-26 | | Release date: | 2007-01-16 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Acidic pH-induced conformational changes in amyloidogenic mutant transthyretin
J.Mol.Biol., 366, 2007
|
|
3CL7
 
 | | Crystal structure of Puue Allantoinase in complex with Hydantoin | | Descriptor: | Puue Allantoinase, imidazolidine-2,4-dione | | Authors: | Ramazzina, I, Cendron, L, Folli, C, Berni, R, Monteverdi, D, Zanotti, G, Percudani, R. | | Deposit date: | 2008-03-18 | | Release date: | 2008-06-10 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Logical identification of an allantoinase analog (puuE) recruited from polysaccharide deacetylases
J.Biol.Chem., 283, 2008
|
|
3CL8
 
 | | Crystal structure of Puue Allantoinase complexed with ACA | | Descriptor: | 5-amino-1H-imidazole-4-carboxamide, Puue Allantoinase | | Authors: | Ramazzina, I, Cendron, L, Folli, C, Berni, R, Monteverdi, D, Zanotti, G, Percudani, R. | | Deposit date: | 2008-03-18 | | Release date: | 2008-06-10 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Logical identification of an allantoinase analog (puuE) recruited from polysaccharide deacetylases
J.Biol.Chem., 283, 2008
|
|
3CL6
 
 | | Crystal structure of Puue Allantoinase | | Descriptor: | Puue allantoinase | | Authors: | Ramazzina, I, Cendron, L, Folli, C, Berni, R, Monteverdi, D, Zanotti, G, Percudani, R. | | Deposit date: | 2008-03-18 | | Release date: | 2008-06-10 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Logical identification of an allantoinase analog (puuE) recruited from polysaccharide deacetylases
J.Biol.Chem., 283, 2008
|
|
