6F2G
 
 | | Bacterial asc transporter crystal structure in open to in conformation | | Descriptor: | Nanobody 74, Putative amino acid/polyamine transport protein, ZINC ION | | Authors: | Fort, J, Errasti-Murugarren, E, Carpena, X, Palacin, M, Fita, I. | | Deposit date: | 2017-11-24 | | Release date: | 2019-04-24 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.92 Å) | | Cite: | L amino acid transporter structure and molecular bases for the asymmetry of substrate interaction.
Nat Commun, 10, 2019
|
|
6F2W
 
 | | Bacterial asc transporter crystal structure in open to in conformation | | Descriptor: | ALPHA-AMINOISOBUTYRIC ACID, Nanobody 74, Putative amino acid/polyamine transport protein, ... | | Authors: | Fort, J, Errasti-Murugarren, E, Carpena, X, Palacin, M, Fita, I. | | Deposit date: | 2017-11-27 | | Release date: | 2019-04-24 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | L amino acid transporter structure and molecular bases for the asymmetry of substrate interaction.
Nat Commun, 10, 2019
|
|
1A3R
 
 | |
2V5H
 
 | | Controlling the storage of nitrogen as arginine: the complex of PII and acetylglutamate kinase from Synechococcus elongatus PCC 7942 | | Descriptor: | ACETYLGLUTAMATE KINASE, CHLORIDE ION, GLYCEROL, ... | | Authors: | Llacer, J.L, Marco-Marin, C, Gil-Ortiz, F, Fita, I, Rubio, V. | | Deposit date: | 2007-07-04 | | Release date: | 2007-10-16 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | The Crystal Structure of the Complex of Pii and Acetylglutamate Kinase Reveals How Pii Controls the Storage of Nitrogen as Arginine.
Proc.Natl.Acad.Sci.USA, 104, 2007
|
|
1YDX
 
 | | Crystal structure of Type-I restriction-modification system S subunit from M. genitalium | | Descriptor: | CHLORIDE ION, type I restriction enzyme specificity protein MG438 | | Authors: | Machado, B, Quijada, O, Pinol, J, Fita, I, Querol, E, Carpena, X. | | Deposit date: | 2004-12-27 | | Release date: | 2005-08-23 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of a Putative Type I Restriction-Modification S Subunit from Mycoplasma genitalium
J.Mol.Biol., 351, 2005
|
|
4AB7
 
 | | Crystal structure of a tetrameric acetylglutamate kinase from Saccharomyces cerevisiae complexed with its substrate N- acetylglutamate | | Descriptor: | N-ACETYL-L-GLUTAMATE, PROTEIN ARG5,6, MITOCHONDRIAL | | Authors: | de Cima, S, Gil-Ortiz, F, Crabeel, M, Fita, I, Rubio, V. | | Deposit date: | 2011-12-07 | | Release date: | 2012-05-02 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Insight on an Arginine Synthesis Metabolon from the Tetrameric Structure of Yeast Acetylglutamate Kinase
Plos One, 7, 2012
|
|
1YE9
 
 | | Crystal structure of proteolytically truncated catalase HPII from E. coli | | Descriptor: | CIS-HEME D HYDROXYCHLORIN GAMMA-SPIROLACTONE, catalase HPII | | Authors: | Loewen, P.C, Chelikani, P, Carpena, X, Fita, I, Perez-Luque, R, Donald, L.J, Switala, J, Duckworth, H.W. | | Deposit date: | 2004-12-28 | | Release date: | 2005-04-26 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Characterization of a Large Subunit Catalase Truncated by Proteolytic Cleavage(,)
Biochemistry, 44, 2005
|
|
1BBD
 
 | |
1CF9
 
 | |
5DOU
 
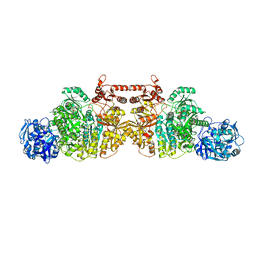 | | Crystal Structure of Human Carbamoyl phosphate synthetase I (CPS1), ligand-bound form | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5'-DIPHOSPHATE, CHLORIDE ION, ... | | Authors: | de Cima, S, Polo, L.M, Fita, I, Rubio, V. | | Deposit date: | 2015-09-11 | | Release date: | 2015-12-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of human carbamoyl phosphate synthetase: deciphering the on/off switch of human ureagenesis.
Sci Rep, 5, 2015
|
|
5DOT
 
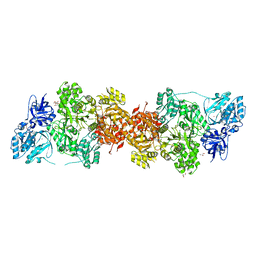 | | Crystal Structure of Human Carbamoyl phosphate synthetase I (CPS1), apo form | | Descriptor: | 1,2-ETHANEDIOL, Carbamoyl-phosphate synthase [ammonia], mitochondrial, ... | | Authors: | Polo, L.M, de Cima, S, Fita, I, Rubio, V. | | Deposit date: | 2015-09-11 | | Release date: | 2015-12-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of human carbamoyl phosphate synthetase: deciphering the on/off switch of human ureagenesis.
Sci Rep, 5, 2015
|
|
3F9P
 
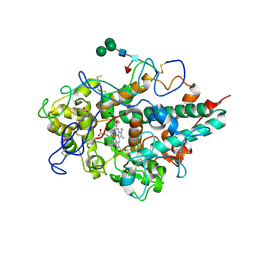 | | Crystal structure of myeloperoxidase from human leukocytes | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, CALCIUM ION, ... | | Authors: | Carpena, X, Fita, I, Obinger, C. | | Deposit date: | 2008-11-14 | | Release date: | 2009-07-14 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.93 Å) | | Cite: | Essential role of proximal histidine-asparagine interaction in Mammalian peroxidases.
J.Biol.Chem., 284, 2009
|
|
3DPR
 
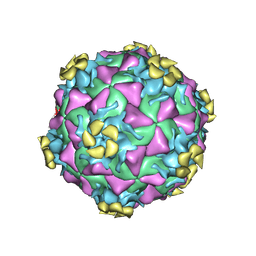 | | Human rhinovirus 2 bound to a concatamer of the VLDL receptor module V3 | | Descriptor: | CALCIUM ION, LAURIC ACID, LDL-receptor class A 3, ... | | Authors: | Querol-Audi, J, Pous, J, Fita, I, Verdaguer, N. | | Deposit date: | 2008-07-09 | | Release date: | 2009-04-07 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Minor group human rhinovirus-receptor interactions: geometry of multimodular attachment and basis of recognition
Febs Lett., 583, 2009
|
|
4DCZ
 
 | |
5CJH
 
 | | Crystal Structure of Eukaryotic Oxoiron MagKatG2 at pH 8.5 | | Descriptor: | Catalase-peroxidase 2, HYDROXIDE ION, Peroxidized Heme | | Authors: | Gasselhuber, B, Obinger, C, Fita, I, Carpena, X. | | Deposit date: | 2015-07-14 | | Release date: | 2015-09-02 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Eukaryotic Catalase-Peroxidase: The Role of the Trp-Tyr-Met Adduct in Protein Stability, Substrate Accessibility, and Catalysis of Hydrogen Peroxide Dismutation.
Biochemistry, 54, 2015
|
|
2O4G
 
 | | Structure of TREX1 in complex with a nucleotide | | Descriptor: | MAGNESIUM ION, THYMIDINE-5'-PHOSPHATE, Three prime repair exonuclease 1 | | Authors: | Brucet, M, Macias, M.J, Fita, I, Celada, A. | | Deposit date: | 2006-12-04 | | Release date: | 2007-03-13 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structure of the dimeric exonuclease TREX1 in complex with DNA displays a proline-rich binding site for WW Domains.
J.Biol.Chem., 282, 2007
|
|
2OBY
 
 | | Crystal structure of Human P53 inducible oxidoreductase (TP53I3,PIG3) | | Descriptor: | NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Putative quinone oxidoreductase | | Authors: | Porte, S, Valencia, E, Farres, J, Fita, I, Pike, A.C.W, Shafqat, N, Debreczeni, J, Johansson, C, Haroniti, A, Gileadi, O, Arrowsmith, C.H, Edwards, A, Weigelt, J, Sundstrom, M, von Delft, F, Oppermann, U, Pares, X. | | Deposit date: | 2006-12-20 | | Release date: | 2007-01-30 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of human P53 inducible oxidoreductase (TP53I3, PIG3)
To be Published
|
|
3UPL
 
 | | Crystal structure of the Brucella abortus enzyme catalyzing the first committed step of the methylerythritol 4-phosphate pathway. | | Descriptor: | GLYCEROL, MAGNESIUM ION, Oxidoreductase | | Authors: | Calisto, B.M, Perez-Gil, J, Fita, I, Rodriguez-Concepcion, M. | | Deposit date: | 2011-11-18 | | Release date: | 2012-03-28 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of Brucella abortus deoxyxylulose-5-phosphate reductoisomerase-like (DRL) enzyme involved in isoprenoid biosynthesis.
J.Biol.Chem., 287, 2012
|
|
1ZUA
 
 | | Crystal Structure Of AKR1B10 Complexed With NADP+ And Tolrestat | | Descriptor: | Aldo-keto reductase family 1 member B10, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, TOLRESTAT | | Authors: | Gallego, O, Ruiz, F.X, Ardevol, A, Dominguez, M, Alvarez, R, de Lera, A.R, Rovira, C, Farres, J, Fita, I, Pares, X. | | Deposit date: | 2005-05-30 | | Release date: | 2006-11-07 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Structural basis for the high all-trans-retinaldehyde reductase activity of the tumor marker AKR1B10.
Proc.Natl.Acad.Sci.USA, 104, 2007
|
|
2PMP
 
 | | Structure of 2C-methyl-D-erythritol 2,4-cyclodiphosphate synthase from the isoprenoid biosynthetic pathway of Arabidopsis thaliana | | Descriptor: | 2-C-methyl-D-erythritol 2,4-cyclodiphosphate synthase, CHLORIDE ION, CYTIDINE-5'-MONOPHOSPHATE, ... | | Authors: | Calisto, B.M, Perez-Gil, J, Querol-Audi, J, Fita, I, Imperial, S. | | Deposit date: | 2007-04-23 | | Release date: | 2007-09-18 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Biosynthesis of isoprenoids in plants: Structure of the 2C-methyl-D-erithrytol 2,4-cyclodiphosphate synthase from Arabidopsis thaliana. Comparison with the bacterial enzymes.
Protein Sci., 16, 2007
|
|
3GF5
 
 | | Crystal structure of the P21 R1-R7 N-terminal domain of murine MVP | | Descriptor: | GLYCEROL, Major vault protein | | Authors: | Querol-Audi, J, Casanas, A, Luque, D, Caston, J.R, Fita, I, Verdaguer, N. | | Deposit date: | 2009-02-26 | | Release date: | 2009-11-10 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The mechanism of vault opening from the high resolution structure of the N-terminal repeats of MVP
Embo J., 28, 2009
|
|
1GGJ
 
 | | CRYSTAL STRUCTURE OF CATALASE HPII FROM ESCHERICHIA COLI, ASN201ALA VARIANT. | | Descriptor: | CATALASE HPII, CIS-HEME D HYDROXYCHLORIN GAMMA-SPIROLACTONE | | Authors: | Melik-Adamyan, W.R, Bravo, J, Carpena, X, Switala, J, Mate, M.J, Fita, I, Loewen, P.C. | | Deposit date: | 2000-08-21 | | Release date: | 2000-08-30 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Substrate flow in catalases deduced from the crystal structures of active site variants of HPII from Escherichia coli.
Proteins, 44, 2001
|
|
1GGE
 
 | | CRYSTAL STRUCTURE OF CATALASE HPII FROM ESCHERICHIA COLI, NATIVE STRUCTURE AT 1.9 A RESOLUTION. | | Descriptor: | CIS-HEME D HYDROXYCHLORIN GAMMA-SPIROLACTONE, PROTEIN (CATALASE HPII) | | Authors: | Melik-Adamyan, W.R, Bravo, J, Carpena, X, Switala, J, Mate, M.J, Fita, I, Loewen, P.C. | | Deposit date: | 2000-08-16 | | Release date: | 2000-08-30 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Substrate flow in catalases deduced from the crystal structures of active site variants of HPII from Escherichia coli.
Proteins, 44, 2001
|
|
3B6O
 
 | | Structure of TREX1 in complex with a nucleotide and an inhibitor ion (lithium) | | Descriptor: | LITHIUM ION, THYMIDINE-5'-PHOSPHATE, Three prime repair exonuclease 1 | | Authors: | Brucet, M, Querol-Audi, J, Fita, I, Celada, A. | | Deposit date: | 2007-10-29 | | Release date: | 2008-09-23 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural and biochemical studies of TREX1 inhibition by metals. Identification of a new active histidine conserved in DEDDh exonucleases.
Protein Sci., 17, 2008
|
|
3B6P
 
 | | Structure of TREX1 in complex with a nucleotide and inhibitor ions (sodium and zinc) | | Descriptor: | SODIUM ION, THYMIDINE-5'-PHOSPHATE, Three prime repair exonuclease 1, ... | | Authors: | Brucet, M, Querol-Audi, J, Fita, I, Celada, A. | | Deposit date: | 2007-10-29 | | Release date: | 2008-09-23 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural and biochemical studies of TREX1 inhibition by metals. Identification of a new active histidine conserved in DEDDh exonucleases.
Protein Sci., 17, 2008
|
|
