6S7W
 
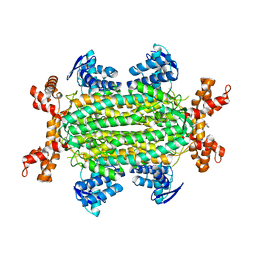 | | Fumarate hydratase of Mycobacterium tuberculosis in complex with formate and allosteric modulator N-(5-(Azepan-1-ylsulfonyl)-2-methoxyphenyl)-2-(quinolin-4-yl)acetamide | | Descriptor: | FORMIC ACID, Fumarate hydratase class II, MAGNESIUM ION, ... | | Authors: | Whitehouse, A.J, Libardo, M.D, Kasbekar, M, Brear, P, Fischer, G, Thomas, C.J, Barry, C.E, Boshoff, H.I, Coyne, A.G, Abell, C. | | Deposit date: | 2019-07-07 | | Release date: | 2019-09-25 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | Targeting of Fumarate Hydratase fromMycobacterium tuberculosisUsing Allosteric Inhibitors with a Dimeric-Binding Mode.
J.Med.Chem., 62, 2019
|
|
4CXK
 
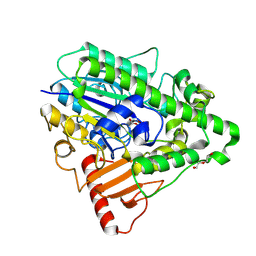 | | G9 mutant of PAS, arylsulfatase from Pseudomonas Aeruginosa | | Descriptor: | ARYLSULFATASE, CALCIUM ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Miton, C.M, Jonas, S, Mohammed, M.F, Fischer, G, Loo, B.v, Kintses, B, Hyvonen, M, Tokuriki, N, Hollfelder, F. | | Deposit date: | 2014-04-07 | | Release date: | 2015-04-22 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Evolutionary repurposing of a sulfatase: A new Michaelis complex leads to efficient transition state charge offset.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
4CYS
 
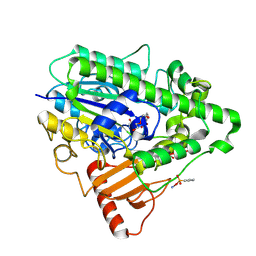 | | G6 mutant of PAS, arylsulfatase from Pseudomonas Aeruginosa, in complex with Phenylphosphonic acid | | Descriptor: | AMMONIUM ION, ARYLSULFATASE, CALCIUM ION, ... | | Authors: | Miton, C.M, Jonas, S, Mohammed, M.F, Fischer, G, Loo, B.v, Kintses, B, Hyvonen, M, Tokuriki, N, Hollfelder, F. | | Deposit date: | 2014-04-14 | | Release date: | 2015-04-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Evolutionary repurposing of a sulfatase: A new Michaelis complex leads to efficient transition state charge offset.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
4CXU
 
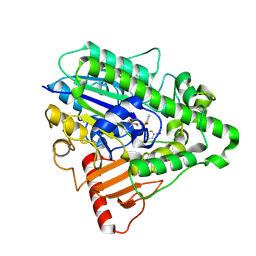 | | G4 mutant of PAS, arylsulfatase from Pseudomonas Aeruginosa, in complex with 3-Br-Phenolphenylphosphonate | | Descriptor: | 3-bromophenyl hydrogen (S)-phenylphosphonate, ARYLSULFATASE, CALCIUM ION | | Authors: | Miton, C.M, Jonas, S, Mohammed, M.F, Fischer, G, Loo, B.v, Kintses, B, Hyvonen, M, Tokuriki, N, Hollfelder, F. | | Deposit date: | 2014-04-08 | | Release date: | 2015-04-22 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Evolutionary repurposing of a sulfatase: A new Michaelis complex leads to efficient transition state charge offset.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
5OOV
 
 | | Designed Ankyrin Repeat Protein (DARPin) ETVD-1 in complex with Lysozyme | | Descriptor: | DARPin ETVD-1, Lysozyme C | | Authors: | Houlihan, G, Fischer, G, Hogan, B.J, Edmond, S, Huovinen, T.T.K, Hollfelder, F, Hyvonen, M. | | Deposit date: | 2017-08-08 | | Release date: | 2018-08-29 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.365 Å) | | Cite: | Designed Ankyrin Repeat Protein (DARPin) ETVD-1 in complex with Lysozyme
To be published
|
|
2F2D
 
 | | Solution structure of the FK506-binding domain of human FKBP38 | | Descriptor: | 38 kDa FK-506 binding protein homolog, FKBP38 | | Authors: | Maestre-Martinez, M, Edlich, F, Jarczowski, F, Weiwad, M, Fischer, G, Luecke, C. | | Deposit date: | 2005-11-16 | | Release date: | 2006-05-02 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of the FK506-Binding Domain of Human FKBP38
J.BIOMOL.NMR, 34, 2006
|
|
4JA9
 
 | | Rat PP5 apo | | Descriptor: | MAGNESIUM ION, Serine/threonine-protein phosphatase 5 | | Authors: | Haslbeck, V, Helmuth, M, Alte, F, Popowicz, G, Schmidt, W, Weiwad, M, Fischer, G, Gemmecker, G, Sattler, M, Striggow, F, Groll, M, Richter, K. | | Deposit date: | 2013-02-18 | | Release date: | 2014-02-19 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Selective activators of protein phosphatase 5 target the auto-inhibitory mechanism.
Biosci.Rep., 35, 2015
|
|
4JA7
 
 | | Rat PP5 co-crystallized with P5SA-2 | | Descriptor: | MAGNESIUM ION, Serine/threonine-protein phosphatase 5 | | Authors: | Haslbeck, V, Helmuth, M, Alte, F, Popowicz, G, Schmidt, W, Weiwad, M, Fischer, G, Gemmecker, G, Sattler, M, Striggow, F, Groll, M, Richter, K. | | Deposit date: | 2013-02-18 | | Release date: | 2014-02-19 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Selective activators of protein phosphatase 5 target the auto-inhibitory mechanism.
Biosci.Rep., 35, 2015
|
|
5NXS
 
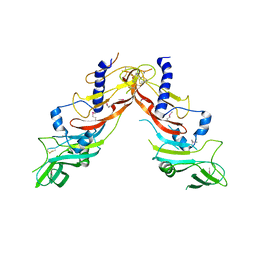 | |
5L8V
 
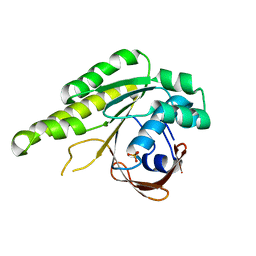 | | Apo-structure of humanised RadA-mutant humRadA4 | | Descriptor: | DNA repair and recombination protein RadA, PHOSPHATE ION | | Authors: | Marsh, M, Fischer, G, Moschetti, T, Sharpe, T, Scott, D, Morgan, M, Ng, H, Skidmore, J, Venkitaraman, A, Abell, C, Blundell, T.L, Hyvonen, M. | | Deposit date: | 2016-06-08 | | Release date: | 2016-10-19 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Engineering Archeal Surrogate Systems for the Development of Protein-Protein Interaction Inhibitors against Human RAD51.
J.Mol.Biol., 428, 2016
|
|
5LB4
 
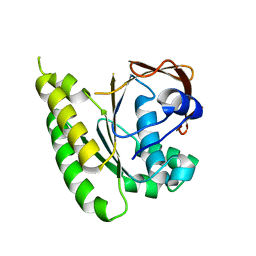 | | Apo-structure of humanised RadA-mutant humRadA14 | | Descriptor: | DNA repair and recombination protein RadA | | Authors: | Marsh, M, Fischer, G, Moschetti, T, Sharpe, T, Scott, D, Morgan, M, Ng, H, Skidmore, J, Venkitaraman, A, Abell, C, Blundell, T.L, Hyvonen, M. | | Deposit date: | 2016-06-15 | | Release date: | 2016-10-19 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Engineering Archeal Surrogate Systems for the Development of Protein-Protein Interaction Inhibitors against Human RAD51.
J.Mol.Biol., 428, 2016
|
|
5LBI
 
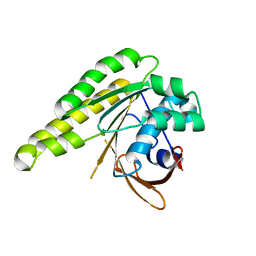 | | Apo-structure of humanised RadA-mutant humRadA3 | | Descriptor: | CALCIUM ION, DNA repair and recombination protein RadA | | Authors: | Marsh, M, Fischer, G, Moschetti, T, Sharpe, T, Scott, D, Morgan, M, Ng, H, Skidmore, J, Venkitaraman, A, Abell, C, Blundell, T.L, Hyvonen, M. | | Deposit date: | 2016-06-16 | | Release date: | 2016-10-26 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Engineering Archeal Surrogate Systems for the Development of Protein-Protein Interaction Inhibitors against Human RAD51.
J.Mol.Biol., 428, 2016
|
|
5LB2
 
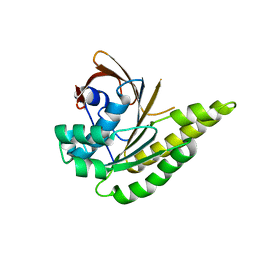 | | Apo-structure of humanised RadA-mutant humRadA2 | | Descriptor: | DNA repair and recombination protein RadA | | Authors: | Marsh, M, Fischer, G, Moschetti, T, Sharpe, T, Scott, D, Morgan, M, Ng, H, Skidmore, J, Venkitaraman, A, Abell, C, Blundell, T.L, Hyvonen, M. | | Deposit date: | 2016-06-15 | | Release date: | 2016-10-19 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Engineering Archeal Surrogate Systems for the Development of Protein-Protein Interaction Inhibitors against Human RAD51.
J.Mol.Biol., 428, 2016
|
|
2UZ5
 
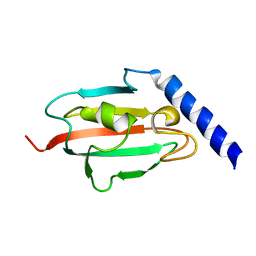 | | Solution structure of the fkbp-domain of Legionella pneumophila Mip | | Descriptor: | MACROPHAGE INFECTIVITY POTENTIATOR | | Authors: | Ceymann, A, Horstmann, M, Ehses, P, Schweimer, K, Steinert, M, Kamphausen, T, Fischer, G, Hacker, J, Rosch, P, Faber, C. | | Deposit date: | 2007-04-25 | | Release date: | 2007-06-12 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Domain Motions of the Mip Protein from Legionella Pneumophila
Biochemistry, 45, 2006
|
|
5OOY
 
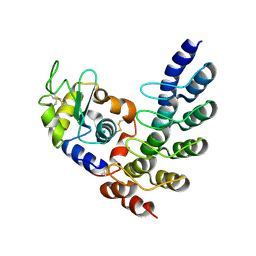 | | Designed Ankyrin Repeat Protein (DARPin) VHAH-1 in complex with Lysozyme | | Descriptor: | DARPin VHAH-1, Lysozyme C | | Authors: | Hogan, B.J, Fischer, G, Houlihan, G, Edmond, S, Huovinen, T.T.K, Hollfelder, F, Hyvonen, M. | | Deposit date: | 2017-08-09 | | Release date: | 2018-08-29 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Designed Ankyrin Repeat Protein (DARPin) VHAH-1 in complex with Lysozyme
To be published
|
|
6SFR
 
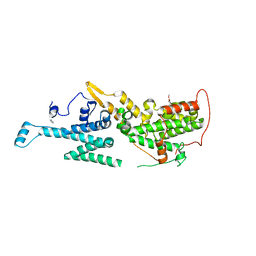 | | SOS1 in Complex with Inhibitor BI-68BS | | Descriptor: | 6,7-dimethoxy-~{N}-[(1~{R})-1-phenylethyl]quinazolin-4-amine, IMIDAZOLE, Son of sevenless homolog 1 | | Authors: | Kessler, D, Fischer, G, Ramharter, J. | | Deposit date: | 2019-08-02 | | Release date: | 2020-08-26 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.917 Å) | | Cite: | BI-3406, a Potent and Selective SOS1-KRAS Interaction Inhibitor, Is Effective in KRAS-Driven Cancers through Combined MEK Inhibition.
Cancer Discov, 11, 2021
|
|
6SCM
 
 | | SOS1 in Complex with Inhibitor BI-3406 | | Descriptor: | 1,2-ETHANEDIOL, IMIDAZOLE, Son of sevenless homolog 1, ... | | Authors: | Kessler, D, Fischer, G, Ramharter, J. | | Deposit date: | 2019-07-24 | | Release date: | 2020-08-26 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.866 Å) | | Cite: | BI-3406, a Potent and Selective SOS1-KRAS Interaction Inhibitor, Is Effective in KRAS-Driven Cancers through Combined MEK Inhibition.
Cancer Discov, 11, 2021
|
|
2VCD
 
 | | Solution structure of the FKBP-domain of Legionella pneumophila Mip in complex with rapamycin | | Descriptor: | Outer membrane protein MIP, RAPAMYCIN IMMUNOSUPPRESSANT DRUG | | Authors: | Ceymann, A, Horstmann, M, Ehses, P, Schweimer, K, Paschke, A.-K, Fischer, G, Roesch, P, Faber, C. | | Deposit date: | 2007-09-20 | | Release date: | 2008-09-02 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the Legionella pneumophila Mip-rapamycin complex.
BMC Struct. Biol., 8, 2008
|
|
