4URT
 
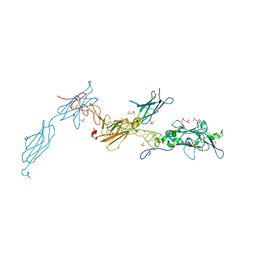 | | The crystal structure of a fragment of netrin-1 in complex with FN5- FN6 of DCC | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Finci, L.I, Krueger, N, Sun, X, Zhang, J, Chegkazi, M, Wu, Y, Schenk, G, Mertens, H.D.T, Svergun, D.I, Zhang, Y, Wang, J.-h, Meijers, R. | | Deposit date: | 2014-07-02 | | Release date: | 2014-09-10 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | The Crystal Structure of Netrin-1 in Complex with Dcc Reveals the Bi-Functionality of Netrin-1 as a Guidance Cue
Neuron, 83, 2014
|
|
1QWA
 
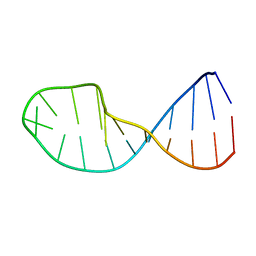 | | NMR structure of 5'-r(GGAUGCCUCCCGAGUGCAUCC): an RNA hairpin derived from the mouse 5'ETS that binds nucleolin RBD12. | | Descriptor: | 18S ribosomal RNA, 5'ETS | | Authors: | Finger, L.D, Trantirek, L, Johansson, C, Feigon, J. | | Deposit date: | 2003-09-01 | | Release date: | 2003-11-25 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution Strucutres of Stem-loop RNAs that Bind to the Two N-terminal RNA Binding Domains of Nucleolin
Nucleic Acids Res., 31, 2003
|
|
1QWB
 
 | |
8U1M
 
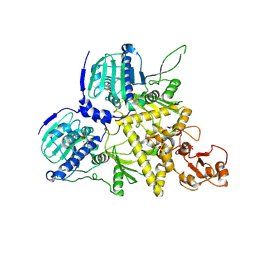 | |
8U1N
 
 | |
5WH9
 
 | |
8U1L
 
 | |
5ZYA
 
 | | SF3b spliceosomal complex bound to E7107 | | Descriptor: | PHD finger-like domain-containing protein 5A, POTASSIUM ION, Splicing factor 3B subunit 1, ... | | Authors: | Finci, L.I, Larsen, N.A. | | Deposit date: | 2018-05-23 | | Release date: | 2018-06-20 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.95 Å) | | Cite: | The cryo-EM structure of the SF3b spliceosome complex bound to a splicing modulator reveals a pre-mRNA substrate competitive mechanism of action
Genes Dev., 32, 2018
|
|
3FM9
 
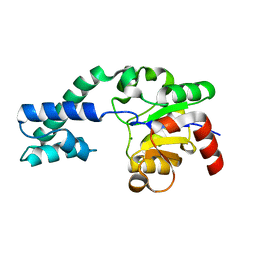 | | Analysis of the Structural Determinants Underlying Discrimination between Substrate and Solvent in beta-Phosphoglucomutase Catalysis | | Descriptor: | Beta-phosphoglucomutase, MAGNESIUM ION | | Authors: | Finci, L, Lahiri, S, Peisach, E, Allen, K.N. | | Deposit date: | 2008-12-19 | | Release date: | 2009-06-09 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Analysis of the structural determinants underlying discrimination between substrate and solvent in beta-phosphoglucomutase catalysis.
Biochemistry, 48, 2009
|
|
6STR
 
 | | Three dimensional structure of the giant reed (Arundodonax) lectin (ADL) complex with N,N'-Diacetylchitobiose; 60 seconds soaking | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Arundo donax Lectin (ADL), GLYCEROL | | Authors: | Perduca, M, Monaco, H.L, Bovi, M, Destefanis, L, Nadali, D, Fin, L, Carrizo, M.E. | | Deposit date: | 2019-09-11 | | Release date: | 2021-07-14 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Three-dimensional structure and properties of the giant reed (Arundo donax) lectin (ADL).
Glycobiology, 2021
|
|
6STQ
 
 | | Three dimensional structure of the giant reed (Arundodonax) lectin (ADL) complex with N,N'-Diacetylchitobiose; 30 seconds soaking | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Arundo donax Lectin (ADL), GLYCEROL | | Authors: | Perduca, M, Monaco, H.L, Bovi, M, Destefanis, L, Nadali, D, Fin, L, Carrizo, M.E. | | Deposit date: | 2019-09-11 | | Release date: | 2021-07-14 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Three-dimensional structure and properties of the giant reed (Arundo donax) lectin (ADL).
Glycobiology, 2021
|
|
6STN
 
 | | Three dimensional structure of the giant reed (Arundodonax) lectin (ADL) complex with N-Acetyl glucosamine | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Arundo donax Lectin (ADL), GLYCEROL | | Authors: | Perduca, M, Monaco, H.L, Bovi, M, Destefanis, L, Nadali, D, Fin, L, Carrizo, M.E. | | Deposit date: | 2019-09-11 | | Release date: | 2021-07-14 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Three-dimensional structure and properties of the giant reed (Arundo donax) lectin (ADL).
Glycobiology, 2021
|
|
6STM
 
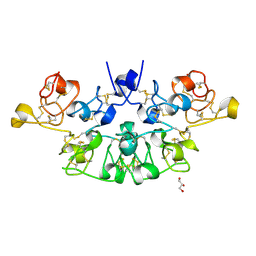 | | Three dimensional structure of the giant reed (Arundodonax) lectin (ADL) | | Descriptor: | Arundo donax Lectin (ADL), GLYCEROL | | Authors: | Perduca, M, Monaco, H.L, Bovi, M, Destefanis, L, Nadali, D, Fin, L, Carrizo, M.E. | | Deposit date: | 2019-09-11 | | Release date: | 2021-07-14 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Three-dimensional structure and properties of the giant reed (Arundo donax) lectin (ADL).
Glycobiology, 2021
|
|
6STP
 
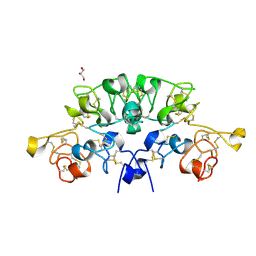 | | Three dimensional structure of the giant reed (Arundodonax) lectin (ADL) complex with sialic acid | | Descriptor: | Arundo donax Lectin (ADL), GLYCEROL, N-acetyl-alpha-neuraminic acid | | Authors: | Perduca, M, Monaco, H.L, Bovi, M, Destefanis, L, Nadali, D, Fin, L, Carrizo, M.E. | | Deposit date: | 2019-09-11 | | Release date: | 2021-07-14 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Three-dimensional structure and properties of the giant reed (Arundo donax) lectin (ADL).
Glycobiology, 2021
|
|
6STO
 
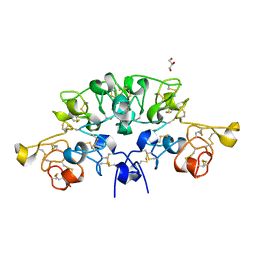 | | Three dimensional structure of the giant reed (Arundodonax) lectin (ADL) complex with N-Acetyl lactosamine | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Arundo donax Lectin (ADL), GLYCEROL, ... | | Authors: | Perduca, M, Monaco, H.L, Bovi, M, Destefanis, L, Nadali, D, Fin, L, Carrizo, M.E. | | Deposit date: | 2019-09-11 | | Release date: | 2021-07-14 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Three-dimensional structure and properties of the giant reed (Arundo donax) lectin (ADL).
Glycobiology, 2021
|
|
6WXY
 
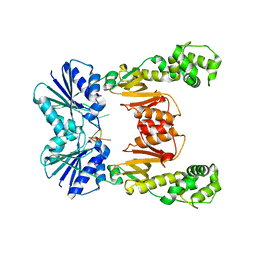 | | crystal structure of cA6-bound Card1 | | Descriptor: | Card1, cA6 | | Authors: | Rostol, J, Xie, W, Patel, D.J, Marraffini, L. | | Deposit date: | 2020-05-12 | | Release date: | 2020-12-30 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The Card1 nuclease provides defence during type III CRISPR immunity.
Nature, 590, 2021
|
|
6WXW
 
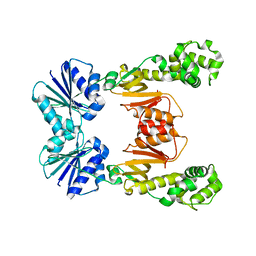 | |
6WXX
 
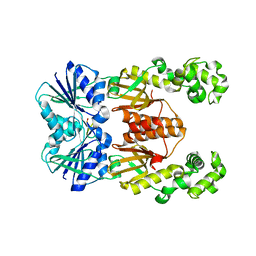 | | crystal structure of cA4-activated Card1 | | Descriptor: | Card1, MANGANESE (II) ION, cA4 | | Authors: | Rostol, J, Xie, W, Patel, D.J, Marraffini, L. | | Deposit date: | 2020-05-12 | | Release date: | 2020-12-30 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The Card1 nuclease provides defence during type III CRISPR immunity.
Nature, 590, 2021
|
|
6XL1
 
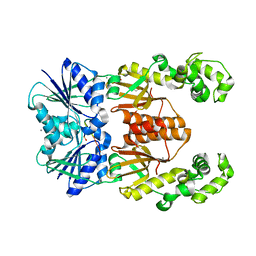 | | crystal structure of cA4-activated Card1(D294N) | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Card1, MANGANESE (II) ION, ... | | Authors: | Rostol, J, Xie, W, Patel, D.J, Marraffini, L. | | Deposit date: | 2020-06-27 | | Release date: | 2020-12-30 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | The Card1 nuclease provides defence during type III CRISPR immunity.
Nature, 590, 2021
|
|
1IE2
 
 | | Solution Structure of an In Vitro Selected RNA which is Sequence Specifically Recognized by RBD12 of Hamster Nucleolin.sNRE (anti) | | Descriptor: | 5'-R(*GP*GP*CP*CP*GP*AP*AP*AP*UP*CP*CP*CP*GP*AP*AP*GP*UP*AP*GP*GP*CP*C)-3' | | Authors: | Bouvet, P, Allain, F.H.-T, Finger, L.D, Dieckmann, T, Feigon, J. | | Deposit date: | 2001-04-05 | | Release date: | 2001-06-20 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Recognition of pre-formed and flexible elements of an RNA stem-loop by nucleolin.
J.Mol.Biol., 309, 2001
|
|
1IE1
 
 | | NMR Solution Structure of an In Vitro Selected RNA which is Sequence Specifically Recognized by Hamster Nucleolin RBD12. | | Descriptor: | 5'-R(*GP*GP*CP*CP*GP*AP*AP*AP*UP*CP*CP*CP*GP*AP*AP*GP*UP*AP*GP*GP*CP*C)-3' | | Authors: | Bouvet, P, Allain, F.H.-T, Finger, L.D, Dieckmann, T, Feigon, J. | | Deposit date: | 2001-04-05 | | Release date: | 2001-06-20 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Recognition of pre-formed and flexible elements of an RNA stem-loop by nucleolin.
J.Mol.Biol., 309, 2001
|
|
1IX2
 
 | | Crystal Structure of Selenomethionine PcoC, a Copper Resistance Protein from Escherichia coli | | Descriptor: | PcoC copper resistance protein | | Authors: | Wernimont, A.K, Huffman, D.L, Finney, L.A, Demeler, B, O'Halloran, T.V, Rosenzweig, A.C. | | Deposit date: | 2002-06-10 | | Release date: | 2002-11-27 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystal structure and dimerization equilibria of PcoC, a methionine-rich copper resistance protein from Escherichia coli
J.BIOL.INORG.CHEM., 8, 2003
|
|
1NA2
 
 | |
6VRB
 
 | | Cryo-EM structure of AcrVIA1-Cas13(crRNA) complex | | Descriptor: | AcrVIA1, CRISPR-associated endoribonuclease Cas13a, RNA (52-MER) | | Authors: | Jia, N, Meeske, A.J, Marraffini, L.A, Patel, D.J. | | Deposit date: | 2020-02-07 | | Release date: | 2020-06-10 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | A phage-encoded anti-CRISPR enables complete evasion of type VI-A CRISPR-Cas immunity.
Science, 369, 2020
|
|
6VRC
 
 | | Cryo-EM structure of Cas13(crRNA) | | Descriptor: | CRISPR-associated endoribonuclease Cas13a, RNA (51-MER) | | Authors: | Jia, N, Meeske, A.J, Marraffini, L.A, Patel, D.J. | | Deposit date: | 2020-02-07 | | Release date: | 2020-06-10 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | A phage-encoded anti-CRISPR enables complete evasion of type VI-A CRISPR-Cas immunity.
Science, 369, 2020
|
|
