1E7K
 
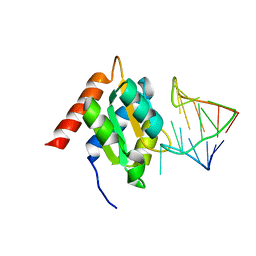 | | Crystal structure of the spliceosomal 15.5kD protein bound to a U4 snRNA fragment | | Descriptor: | 15.5 KD RNA BINDING PROTEIN, RNA (5'-R(*GP*CP*CP*AP*AP*UP*GP*AP*GP*GP*UP*UP*UP* AP*UP*CP*CP*GP*AP*GP*G*C(-3') | | Authors: | Vidovic, I, Nottrott, S, Harthmuth, K, Luhrmann, R, Ficner, R. | | Deposit date: | 2000-08-29 | | Release date: | 2001-02-19 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal Structure of the Spliceosomal 15.5Kd Protein Bound to a U4 Snrna Fragment
Mol.Cell, 6, 2000
|
|
6ZM2
 
 | |
9I2S
 
 | |
1ENU
 
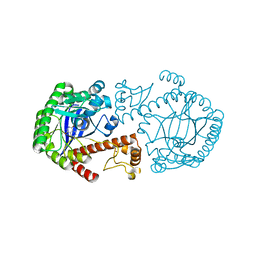 | | A new target for shigellosis: Rational design and crystallographic studies of inhibitors of tRNA-guanine transglycosylase | | Descriptor: | 4-AMINOPHTHALHYDRAZIDE, TRNA-GUANINE TRANSGLYCOSYLASE, ZINC ION | | Authors: | Gradler, U, Gerber, H.D, Goodenough-Lashua, D.M, Garcia, G.A, Ficner, R, Reuter, K, Stubbs, M.T, Klebe, G. | | Deposit date: | 2000-03-21 | | Release date: | 2000-04-19 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | A new target for shigellosis: rational design and crystallographic studies of inhibitors of tRNA-guanine transglycosylase.
J.Mol.Biol., 306, 2001
|
|
4FEE
 
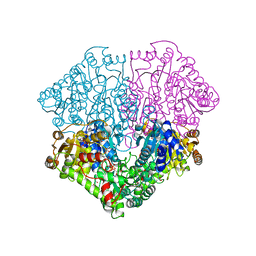 | | High-resolution structure of pyruvate oxidase in complex with reaction intermediate 2-hydroxyethyl-thiamin diphosphate carbanion-enamine, crystal B | | Descriptor: | 2-[(2E)-3-[(4-AMINO-2-METHYLPYRIMIDIN-5-YL)METHYL]-2-(1-HYDROXYETHYLIDENE)-4-METHYL-2,3-DIHYDRO-1,3-THIAZOL-5-YL]ETHYL TRIHYDROGEN DIPHOSPHATE, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, ... | | Authors: | Meyer, D, Neumann, P, Koers, E, Sjuts, H, Luedtke, S, Sheldrick, G.M, Ficner, R, Tittmann, K. | | Deposit date: | 2012-05-30 | | Release date: | 2012-06-20 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.13 Å) | | Cite: | Unexpected tautomeric equilibria of the carbanion-enamine intermediate in pyruvate oxidase highlight unrecognized chemical versatility of thiamin.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4FEG
 
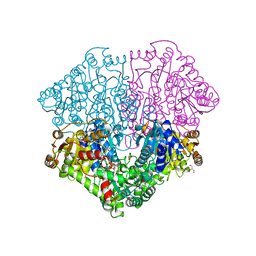 | | High-resolution structure of pyruvate oxidase in complex with reaction intermediate 2-hydroxyethyl-thiamin diphosphate carbanion-enamine, crystal A | | Descriptor: | 2-[(2E)-3-[(4-AMINO-2-METHYLPYRIMIDIN-5-YL)METHYL]-2-(1-HYDROXYETHYLIDENE)-4-METHYL-2,3-DIHYDRO-1,3-THIAZOL-5-YL]ETHYL TRIHYDROGEN DIPHOSPHATE, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, ... | | Authors: | Meyer, D, Neumann, P, Koers, E, Sjuts, H, Luedtke, S, Sheldrick, G.M, Ficner, R, Tittmann, K. | | Deposit date: | 2012-05-30 | | Release date: | 2012-06-20 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.09 Å) | | Cite: | Unexpected tautomeric equilibria of the carbanion-enamine intermediate in pyruvate oxidase highlight unrecognized chemical versatility of thiamin.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
7B51
 
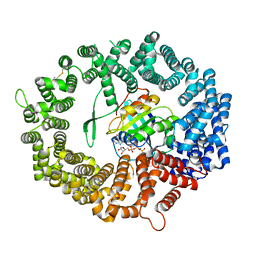 | | Crystal structure of human CRM1 covalently modified by 2-mercaptoethanol at Cys528 | | Descriptor: | BETA-MERCAPTOETHANOL, Exportin-1, GTP-binding nuclear protein Ran, ... | | Authors: | Shaikhqasem, A, Ficner, R. | | Deposit date: | 2020-12-03 | | Release date: | 2021-03-10 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | Crystal structure of human CRM1, covalently modified by 2-mercaptoethanol on Cys528, in complex with RanGTP.
Acta Crystallogr.,Sect.F, 77, 2021
|
|
2QNA
 
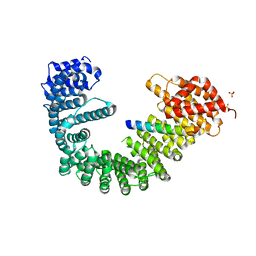 | | Crystal structure of human Importin-beta (127-876) in complex with the IBB-domain of Snurportin1 (1-65) | | Descriptor: | Importin subunit beta-1, SULFATE ION, Snurportin-1 | | Authors: | Wohlwend, D, Strasser, A, Dickmanns, A, Ficner, R. | | Deposit date: | 2007-07-18 | | Release date: | 2008-04-08 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.84 Å) | | Cite: | Structural basis for RanGTP independent entry of spliceosomal U snRNPs into the nucleus.
J.Mol.Biol., 374, 2007
|
|
4FGV
 
 | |
8AF0
 
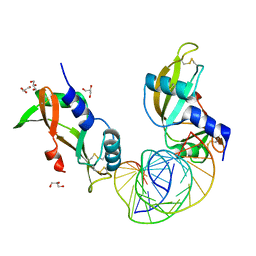 | | Crystal structure of human angiogenin and RNA duplex | | Descriptor: | Angiogenin, GLYCEROL, RNA (5'-R(*GP*CP*CP*CP*GP*CP*CP*UP*GP*UP*CP*AP*CP*GP*CP*GP*GP*GP*C)-3') | | Authors: | Sievers, K, Ficner, R. | | Deposit date: | 2022-07-15 | | Release date: | 2022-09-14 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Structure of angiogenin dimer bound to double-stranded RNA.
Acta Crystallogr.,Sect.F, 78, 2022
|
|
2VCG
 
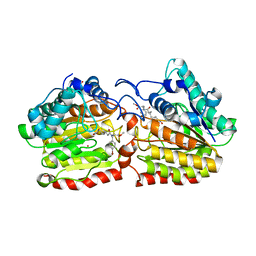 | | Crystal structure of a HDAC-like protein HDAH from Bordetella sp. with the bound inhibitor ST-17 | | Descriptor: | CHLORIDE ION, GLYCEROL, HISTONE DEACETYLASE-LIKE AMIDOHYDROLASE, ... | | Authors: | Dickmanns, A, Strasser, A, Ficner, R. | | Deposit date: | 2007-09-24 | | Release date: | 2008-01-08 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Phenylalanine-Containing Hydroxamic Acids as Selective Inhibitors of Class Iib Histone Deacetylases (Hdacs).
Bioorg.Med.Chem., 16, 2008
|
|
6F2C
 
 | | Methylglyoxal synthase MgsA from Bacillus subtilis | | Descriptor: | CHLORIDE ION, GLYCEROL, Methylglyoxal synthase | | Authors: | Dickmanns, A, Neumann, P, Ficner, R. | | Deposit date: | 2017-11-24 | | Release date: | 2018-03-07 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Structural basis for the regulatory interaction of the methylglyoxal synthase MgsA with the carbon flux regulator Crh inBacillus subtilis.
J. Biol. Chem., 293, 2018
|
|
4TMT
 
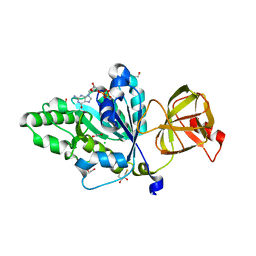 | | Translation initiation factor eIF5B (517-858) mutant D533A from C. thermophilum, bound to GTPgammaS | | Descriptor: | 1,2-ETHANEDIOL, 2,3-DIHYDROXY-1,4-DITHIOBUTANE, 5'-GUANOSINE-DIPHOSPHATE-MONOTHIOPHOSPHATE, ... | | Authors: | Kuhle, B, Ficner, R. | | Deposit date: | 2014-06-02 | | Release date: | 2014-09-24 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | A monovalent cation acts as structural and catalytic cofactor in translational GTPases.
Embo J., 33, 2014
|
|
3GVJ
 
 | | Crystal structure of an endo-neuraminidaseNF mutant | | Descriptor: | Endo-N-acetylneuraminidase, N-acetyl-alpha-neuraminic acid-(2-8)-N-acetyl-alpha-neuraminic acid-(2-8)-N-acetyl-alpha-neuraminic acid-(2-8)-N-acetyl-alpha-neuraminic acid-(2-8)-N-acetyl-beta-neuraminic acid, N-acetyl-alpha-neuraminic acid-(2-8)-N-acetyl-alpha-neuraminic acid-(2-8)-N-acetyl-alpha-neuraminic acid-(2-8)-N-acetyl-beta-neuraminic acid | | Authors: | Schulz, E.C, Dickmanns, A, Ficner, R. | | Deposit date: | 2009-03-31 | | Release date: | 2010-03-02 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Structural basis for the recognition and cleavage of polysialic acid by the bacteriophage K1F tailspike protein EndoNF.
J.Mol.Biol., 397, 2010
|
|
4TMX
 
 | | Translation initiation factor eIF5B (517-858) mutant D533N from C. thermophilum, bound to GTP and sodium | | Descriptor: | 1,2-ETHANEDIOL, ACETIC ACID, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Kuhle, B, Ficner, R. | | Deposit date: | 2014-06-02 | | Release date: | 2014-09-24 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | A monovalent cation acts as structural and catalytic cofactor in translational GTPases.
Embo J., 33, 2014
|
|
2GH6
 
 | | Crystal structure of a HDAC-like protein with 9,9,9-trifluoro-8-oxo-N-phenylnonan amide bound | | Descriptor: | 9,9,9-TRIFLUORO-8-OXO-N-PHENYLNONANAMIDE, Histone deacetylase-like amidohydrolase, POTASSIUM ION, ... | | Authors: | Nielsen, T.K, Hildmann, C, Riester, D, Wegener, D, Schwienhorst, A, Ficner, R. | | Deposit date: | 2006-03-26 | | Release date: | 2007-03-20 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.203 Å) | | Cite: | Complex structure of a bacterial class 2 histone deacetylase homologue with a trifluoromethylketone inhibitor.
Acta Crystallogr.,Sect.F, 63, 2007
|
|
3G8V
 
 | |
4O1K
 
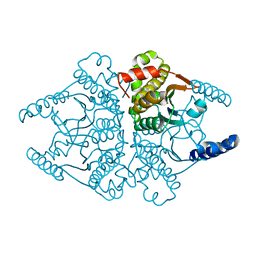 | | Crystal structures of two tetrameric beta-carbonic anhydrases from the filamentous ascomycete Sordaria macrospora. | | Descriptor: | Carbonic anhydrase, ZINC ION | | Authors: | Lehneck, R, Neumann, P, Vullo, D, Elleuche, S, Supuran, C.T, Ficner, R, Poggeler, S. | | Deposit date: | 2013-12-16 | | Release date: | 2014-03-05 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Crystal structures of two tetrameric beta-carbonic anhydrases from the filamentous ascomycete Sordaria macrospora.
Febs J., 281, 2014
|
|
4O1J
 
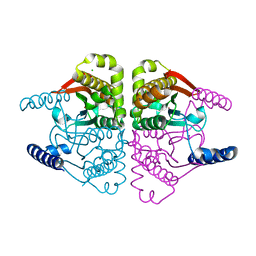 | | Crystal structures of two tetrameric beta-carbonic anhydrases from the filamentous ascomycete Sordaria macrospora. | | Descriptor: | CHLORIDE ION, Carbonic anhydrase, ZINC ION | | Authors: | Lehneck, R, Neumann, P, Vullo, D, Elleuche, S, Supuran, C.T, Ficner, R, Poggeler, S. | | Deposit date: | 2013-12-16 | | Release date: | 2014-03-05 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.695 Å) | | Cite: | Crystal structures of two tetrameric beta-carbonic anhydrases from the filamentous ascomycete Sordaria macrospora.
Febs J., 281, 2014
|
|
6FAC
 
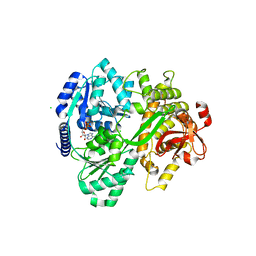 | | CRYSTAL STRUCTURE OF THE DEAH-BOX HELICASE PRP2 IN COMPLEX WITH ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Schmitt, A, Hamann, F, Neumann, P, Ficner, R. | | Deposit date: | 2017-12-15 | | Release date: | 2018-07-11 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal structure of the spliceosomal DEAH-box ATPase Prp2.
Acta Crystallogr D Struct Biol, 74, 2018
|
|
6FAA
 
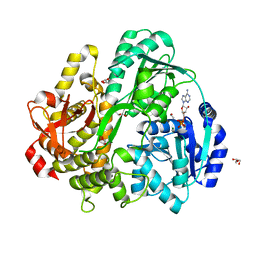 | | CRYSTAL STRUCTURE OF THE DEAH-BOX HELICASE PRP2 IN COMPLEX WITH ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ETHANOL, GLYCEROL, ... | | Authors: | Schmitt, A, Hamann, F, Neumann, P, Ficner, R. | | Deposit date: | 2017-12-15 | | Release date: | 2018-07-11 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Crystal structure of the spliceosomal DEAH-box ATPase Prp2.
Acta Crystallogr D Struct Biol, 74, 2018
|
|
6FA5
 
 | | CRYSTAL STRUCTURE OF THE DEAH-BOX HELICASE PRP2 IN COMPLEX WITH ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, DI(HYDROXYETHYL)ETHER, MAGNESIUM ION, ... | | Authors: | Hamann, F, Schmitt, A, Neumann, P, Ficner, R. | | Deposit date: | 2017-12-15 | | Release date: | 2018-07-11 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.303 Å) | | Cite: | Crystal structure of the spliceosomal DEAH-box ATPase Prp2.
Acta Crystallogr D Struct Biol, 74, 2018
|
|
3GJX
 
 | | Crystal Structure of the Nuclear Export Complex CRM1-Snurportin1-RanGTP | | Descriptor: | CHLORIDE ION, Exportin-1, GTP-binding nuclear protein Ran, ... | | Authors: | Monecke, T, Guettler, T, Neumann, P, Dickmanns, A, Goerlich, D, Ficner, R. | | Deposit date: | 2009-03-09 | | Release date: | 2009-05-26 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of the Nuclear Export Receptor CRM1 in Complex with Snurportin1 and RanGTP.
Science, 2009
|
|
3GUD
 
 | | Crystal structure of a novel intramolecular chaperon | | Descriptor: | BROMIDE ION, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Schulz, E.C, Dickmanns, A, Ficner, R. | | Deposit date: | 2009-03-29 | | Release date: | 2010-02-02 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of an intramolecular chaperone mediating triple-beta-helix folding.
Nat.Struct.Mol.Biol., 17, 2010
|
|
4XRI
 
 | | Crystal Structure of Importin Beta in an Ammonium Sulfate Condition | | Descriptor: | GLYCEROL, Putative uncharacterized protein, SULFATE ION | | Authors: | Tauchert, M.J, Neumann, P, Ficner, R, Dickmanns, A. | | Deposit date: | 2015-01-21 | | Release date: | 2016-01-27 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Impact of the crystallization condition on importin-beta conformation.
Acta Crystallogr D Struct Biol, 72, 2016
|
|
