5LWW
 
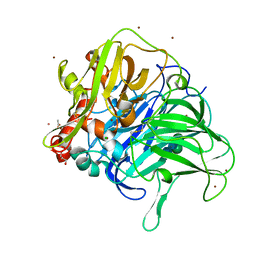 | | Crystal structure of a laccase-like multicopper oxidase McoG from Aspergillus niger bound to zinc | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, ... | | Authors: | Ferraroni, M, Briganti, F, Tamayo-Ramos, J.A, van Berkel, W.J.H, Westphal, A.H. | | Deposit date: | 2016-09-19 | | Release date: | 2017-05-03 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structure and function of Aspergillus niger laccase McoG
Biocatalysis, 2017
|
|
5NEA
 
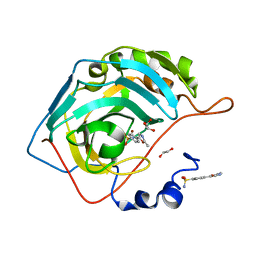 | | Crystal structure of human carbonic anhydrase II in complex with the inhibitor 4-(2-methyl-1,3-oxazol-5-yl)benzene-1-sulfonammide | | Descriptor: | 1,2-ETHANEDIOL, 4-(2-methyl-1,3-oxazol-5-yl)benzenesulfonamide, Carbonic anhydrase 2, ... | | Authors: | Ferraroni, M, Supuran, C.T, Krasavin, M. | | Deposit date: | 2017-03-10 | | Release date: | 2017-12-06 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | 1,3-Oxazole-based selective picomolar inhibitors of cytosolic human carbonic anhydrase II alleviate ocular hypertension in rabbits: Potency is supported by X-ray crystallography of two leads.
Bioorg. Med. Chem., 25, 2017
|
|
8CH4
 
 | | Crystal structure of the ring cleaving dioxygenase 5-nitrosalicylate 1,2-dioxygenase from Bradyrhizobium sp. | | Descriptor: | 5-nitrosalicylic acid 1,2-dioxygenase, FE (III) ION | | Authors: | Ferraroni, M, Stolz, A, Eppinger, E. | | Deposit date: | 2023-02-07 | | Release date: | 2023-07-05 | | Last modified: | 2023-07-12 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of the monocupin ring-cleaving dioxygenase 5-nitrosalicylate 1,2-dioxygenase from Bradyrhizobium sp.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
8B49
 
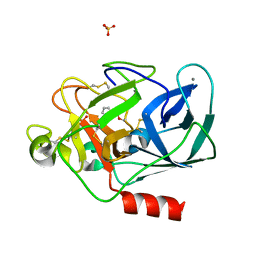 | | STRUCTURE OF PORCINE PANCREATIC ELASTASE BOUND TO A FRAGMENT (m-toluoylcarbonyl group) OF A 5-AZAINDOLE INHIBITOR | | Descriptor: | 1-(3-methylphenyl)carbonylpyrrolo[3,2-c]pyridine-3-carbonitrile, CALCIUM ION, Chymotrypsin-like elastase family member 1, ... | | Authors: | Ferraroni, M, Giovannoni, P, Gerace, A. | | Deposit date: | 2022-09-20 | | Release date: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.19 Å) | | Cite: | X-ray structural study of human neutrophil elastase inhibition with a series of azaindoles, azaindazoles and isoxazolones
J.Mol.Struct., 1274, 2023
|
|
8B53
 
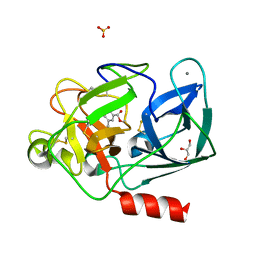 | |
5LWX
 
 | | Crystal structure of the H253D mutant of McoG from Aspergillus niger | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, COPPER (II) ION, ... | | Authors: | Ferraroni, M, Briganti, F, Tamayo-Ramos, J.A, van Berkel, W.J.H, Westphal, A.H. | | Deposit date: | 2016-09-19 | | Release date: | 2017-05-03 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Structure and function of Aspergillus niger laccase McoG
Biocatalysis, 2017
|
|
5LJT
 
 | | Crystal structure of human carbonic anhydrase II in complex with the 4-((1-phenyl-1H-1,2,3-triazol-4-yl)methoxy)benzenesulfonamide inhibitor | | Descriptor: | 4-[[4-[azanyl-bis(oxidanyl)-$l^{4}-sulfanyl]phenoxy]methyl]-1-phenyl-1,2,3-triazole, Carbonic anhydrase 2, GLYCEROL, ... | | Authors: | Ferraroni, M, Supuran, C. | | Deposit date: | 2016-07-19 | | Release date: | 2017-06-21 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Benzenesulfonamides Incorporating Flexible Triazole Moieties Are Highly Effective Carbonic Anhydrase Inhibitors: Synthesis and Kinetic, Crystallographic, Computational, and Intraocular Pressure Lowering Investigations.
J. Med. Chem., 59, 2016
|
|
5NEE
 
 | | Crystal structure of human carbonic anhydrase II in complex with the inhibitor 5-[2-(morpholine-4-carbonyl)1,3-oxazol-5-yl)]thiophene-2-sulfonammide | | Descriptor: | 5-(2-morpholin-4-ylcarbonyl-1,3-oxazol-5-yl)thiophene-2-sulfonamide, Carbonic anhydrase 2, GLYCEROL, ... | | Authors: | Ferraroni, M, Supuran, C.T, Krasavin, M. | | Deposit date: | 2017-03-10 | | Release date: | 2017-12-06 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | 1,3-Oxazole-based selective picomolar inhibitors of cytosolic human carbonic anhydrase II alleviate ocular hypertension in rabbits: Potency is supported by X-ray crystallography of two leads.
Bioorg. Med. Chem., 25, 2017
|
|
6F3C
 
 | | The cytotoxic [Pt(H2bapbpy)] platinum complex interacting with the CGTACG hexamer | | Descriptor: | DNA (5'-D(*GP*TP*AP*CP*G)-3'), MAGNESIUM ION, [Pt(H2bapbpy)] platinum | | Authors: | Ferraroni, M, Bazzicalupi, C, Gratteri, P, Papi, F. | | Deposit date: | 2017-11-28 | | Release date: | 2019-05-29 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Induction of a Four-Way Junction Structure in the DNA Palindromic Hexanucleotide 5'-d(CGTACG)-3' by a Mononuclear Platinum Complex.
Angew.Chem.Int.Ed.Engl., 58, 2019
|
|
4L5K
 
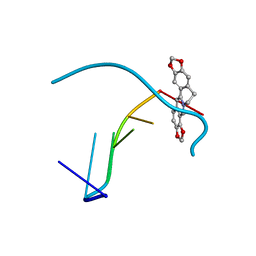 | | Crystal structure of the complex of DNA hexamer d(CGATCG) with Coptisine | | Descriptor: | 6,7-dihydro[1,3]dioxolo[4,5-g][1,3]dioxolo[7,8]isoquino[3,2-a]isoquinolin-5-ium, DNA (5'-D(*CP*GP*AP*TP*CP*G)-3') | | Authors: | Ferraroni, M, Bazzicalupi, C, Gratteri, P. | | Deposit date: | 2013-06-11 | | Release date: | 2014-07-02 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | Crystal structure of the complex of DNA hexamer d(CGATCG) with Coptisine
to be published
|
|
6I0J
 
 | | Crystal structure of human carbonic anhydrase I in complex with the 4-({[4-chloro-3-(trifluoromethyl)phenyl]carbamoyl}amino)phenyl sulfamate inhibitor | | Descriptor: | ACETATE ION, Carbonic anhydrase 1, GLYCEROL, ... | | Authors: | Ferraroni, M, Supuran, C.T, Bozdag, M, Chiapponi, D. | | Deposit date: | 2018-10-26 | | Release date: | 2019-11-20 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Carbonic anhydrase inhibitors based on sorafenib scaffold: Design, synthesis, crystallographic investigation and effects on primary breast cancer cells.
Eur.J.Med.Chem., 182, 2019
|
|
6I0L
 
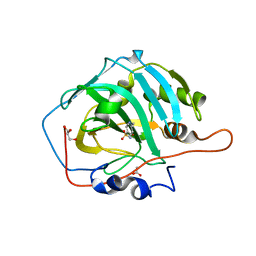 | | Crystal structure of human carbonic anhydrase I in complex with the 1-[4-chloro-3-(trifluoromethyl)phenyl]-3-[2-(4-sulfamoylphenyl)ethyl]urea inhibitor | | Descriptor: | 1-[4-chloranyl-3-(trifluoromethyl)phenyl]-3-[2-(4-sulfamoylphenyl)ethyl]urea, ACETATE ION, Carbonic anhydrase 1, ... | | Authors: | Ferraroni, M, Supuran, C.T, Bozdag, M, Chiapponi, D. | | Deposit date: | 2018-10-26 | | Release date: | 2019-11-20 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Carbonic anhydrase inhibitors based on sorafenib scaffold: Design, synthesis, crystallographic investigation and effects on primary breast cancer cells.
Eur.J.Med.Chem., 182, 2019
|
|
6RHK
 
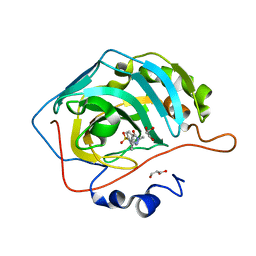 | | The crystal structure of human carbonic anhydrase II in complex with 4-(3-benzylimidazolidine-1-carbonyl)benzenesulfonamide | | Descriptor: | 4-[3-(phenylmethyl)imidazolidin-1-yl]carbonylbenzenesulfonamide, Carbonic anhydrase 2, GLYCEROL, ... | | Authors: | Ferraroni, M, Angeli, A, Supuran, C.T. | | Deposit date: | 2019-04-22 | | Release date: | 2020-05-13 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | Sulfonamides incorporating piperazine bioisosteres as potent human carbonic anhydrase I, II, IV and IX inhibitors.
Bioorg.Chem., 91, 2019
|
|
6S15
 
 | | Pyridine derivative of the natural alkaloid Berberine as Human Telomeric G-quadruplex Binder | | Descriptor: | Berberine, DNA TAGGGTTAGGGT, POTASSIUM ION | | Authors: | Ferraroni, M, Bazzicalupi, C, Gratteri, P, Papi, F. | | Deposit date: | 2019-06-18 | | Release date: | 2020-06-03 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Pyridine Derivative of the Natural Alkaloid Berberine as Human Telomeric G4-DNA Binder: A Solution and Solid-State Study.
Acs Med.Chem.Lett., 11, 2020
|
|
6RG3
 
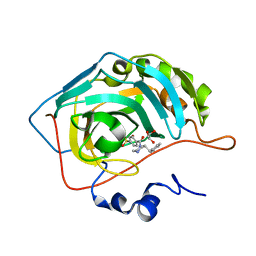 | | Crystal structure of human Carbonic anhydrase II in complex with (R)-4-(2-benzylpiperazin-1-yl)benzenesulfonamide | | Descriptor: | 4-[(2~{R})-2-(phenylmethyl)piperazin-1-yl]carbonylbenzenesulfonamide, Carbonic anhydrase 2, GLYCEROL, ... | | Authors: | Ferraroni, M, Angeli, A, Supuran, C. | | Deposit date: | 2019-04-16 | | Release date: | 2020-05-13 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.32 Å) | | Cite: | Sulfonamides incorporating piperazine bioisosteres as potent human carbonic anhydrase I, II, IV and IX inhibitors.
Bioorg.Chem., 91, 2019
|
|
6RHJ
 
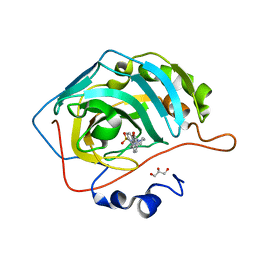 | | Crystal structure of human carbonic anhydrase isozyme II with 4-(4-benzyl-1,4-diazepane-1-carbonyl)benzenesulfonamide | | Descriptor: | 4-[[4-(phenylmethyl)-1,4-diazepan-1-yl]carbonyl]benzenesulfonamide, Carbonic anhydrase 2, GLYCEROL, ... | | Authors: | Ferraroni, M, Angeli, A, Supuran, C.T. | | Deposit date: | 2019-04-21 | | Release date: | 2020-05-13 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | Sulfonamides incorporating piperazine bioisosteres as potent human carbonic anhydrase I, II, IV and IX inhibitors.
Bioorg.Chem., 91, 2019
|
|
6RG4
 
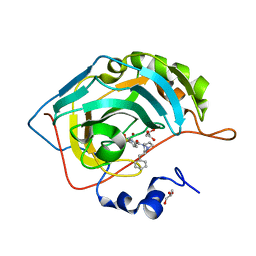 | | Crystal structure of human Carbonic anhydrase II in complex with (R)-4-(2-benzyl-4-methylpiperazin-1-yl)benzenesulfonamide | | Descriptor: | 4-[(3~{S})-4-methyl-3-(phenylmethyl)piperazin-1-yl]carbonylbenzenesulfonamide, Carbonic anhydrase 2, GLYCEROL, ... | | Authors: | Ferraroni, M, Angeli, A, Supuran, C. | | Deposit date: | 2019-04-16 | | Release date: | 2020-05-13 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Sulfonamides incorporating piperazine bioisosteres as potent human carbonic anhydrase I, II, IV and IX inhibitors.
Bioorg.Chem., 91, 2019
|
|
3HGI
 
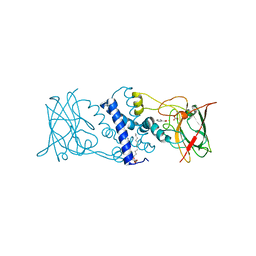 | | Crystal structure of Catechol 1,2-Dioxygenase from the gram-positive Rhodococcus opacus 1CP | | Descriptor: | (4S,7R)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSAN-1-AMINIUM 4-OXIDE, BENZOIC ACID, CARBONATE ION, ... | | Authors: | Ferraroni, M, Matera, I, Briganti, F, Scozzafava, A. | | Deposit date: | 2009-05-14 | | Release date: | 2010-01-12 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Catechol 1,2-dioxygenase from the Gram-positive Rhodococcus opacus 1CP: Quantitative structure/activity relationship and the crystal structures of native enzyme and catechols adducts.
J.Struct.Biol., 170, 2010
|
|
3O6J
 
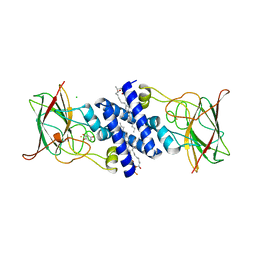 | | Crystal Structure of 4-Chlorocatechol Dioxygenase from Rhodococcus opacus 1CP in complex with hydroxyquinol | | Descriptor: | (2R)-3-(PHOSPHONOOXY)-2-(TETRADECANOYLOXY)PROPYL PALMITATE, CHLORIDE ION, Chlorocatechol 1,2-dioxygenase, ... | | Authors: | Ferraroni, M, Briganti, F, Kolomitseva, M, Golovleva, L. | | Deposit date: | 2010-07-29 | | Release date: | 2011-08-17 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | X-ray structures of 4-chlorocatechol 1,2-dioxygenase adducts with substituted catechols: new perspectives in the molecular basis of intradiol ring cleaving dioxygenases specificity.
J. Struct. Biol., 181, 2013
|
|
5M21
 
 | | Crystal structure of hydroquinone 1,2-dioxygenase from Sphingomonas sp. TTNP3 with 4-hydroxybenzoate bound | | Descriptor: | FE (III) ION, Hydroquinone dioxygenase large subunit, Hydroquinone dioxygenase small subunit, ... | | Authors: | Ferraroni, M, Da Vela, S, Scozzafava, A, Kolvenbach, B, Corvini, P.F.X. | | Deposit date: | 2016-10-11 | | Release date: | 2017-09-27 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | The crystal structures of native hydroquinone 1,2-dioxygenase from Sphingomonas sp. TTNP3 and of substrate and inhibitor complexes.
Biochim. Biophys. Acta, 1865, 2017
|
|
5M22
 
 | | Crystal structure of hydroquinone 1,2-dioxygenase from Sphingomonas sp. TTNP3 | | Descriptor: | FE (III) ION, Hydroquinone dioxygenase large subunit, Hydroquinone dioxygenase small subunit | | Authors: | Ferraroni, M, Da Vela, S, Scozzafava, A, Kolvenbach, B, Corvini, P.F.X. | | Deposit date: | 2016-10-11 | | Release date: | 2017-09-27 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The crystal structures of native hydroquinone 1,2-dioxygenase from Sphingomonas sp. TTNP3 and of substrate and inhibitor complexes.
Biochim. Biophys. Acta, 1865, 2017
|
|
5M4O
 
 | | Crystal structure of hydroquinone 1,2-dioxygenase from Sphingomonas sp. TTNP3 in complex with 4-nitrophenol | | Descriptor: | FE (III) ION, Hydroquinone dioxygenase large subunit, Hydroquinone dioxygenase small subunit, ... | | Authors: | Ferraroni, M, Da Vela, S, Scozzafava, A, Kolvenbach, B, Corvini, P.F.X. | | Deposit date: | 2016-10-18 | | Release date: | 2017-09-27 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The crystal structures of native hydroquinone 1,2-dioxygenase from Sphingomonas sp. TTNP3 and of substrate and inhibitor complexes.
Biochim. Biophys. Acta, 1865, 2017
|
|
5M26
 
 | | Crystal structure of hydroquinone 1,2-dioxygenase from Sphingomonas sp. TTNP3 in complex with methylhydroquinone | | Descriptor: | 2-methylbenzene-1,4-diol, FE (III) ION, Hydroquinone dioxygenase large subunit, ... | | Authors: | Ferraroni, M, Da Vela, S, Scozzafava, A, Kolvenbach, B, Corvini, P.F.X. | | Deposit date: | 2016-10-12 | | Release date: | 2017-09-27 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The crystal structures of native hydroquinone 1,2-dioxygenase from Sphingomonas sp. TTNP3 and of substrate and inhibitor complexes.
Biochim. Biophys. Acta, 1865, 2017
|
|
3O5U
 
 | | Crystal Structure of 4-Chlorocatechol Dioxygenase from Rhodococcus opacus 1CP in complex with protocatechuate | | Descriptor: | (2R)-3-(PHOSPHONOOXY)-2-(TETRADECANOYLOXY)PROPYL PALMITATE, 3,4-DIHYDROXYBENZOIC ACID, CHLORIDE ION, ... | | Authors: | Ferraroni, M, Briganti, F, Kolomitseva, M, Golovleva, L. | | Deposit date: | 2010-07-28 | | Release date: | 2011-08-17 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | X-ray structures of 4-chlorocatechol 1,2-dioxygenase adducts with substituted catechols: new perspectives in the molecular basis of intradiol ring cleaving dioxygenases specificity.
J. Struct. Biol., 181, 2013
|
|
3O6R
 
 | | Crystal Structure of 4-Chlorocatechol Dioxygenase from Rhodococcus opacus 1CP in complex with pyrogallol | | Descriptor: | (2R)-3-(PHOSPHONOOXY)-2-(TETRADECANOYLOXY)PROPYL PALMITATE, BENZENE-1,2,3-TRIOL, Chlorocatechol 1,2-dioxygenase, ... | | Authors: | Ferraroni, M, Briganti, F, Kolomitseva, M, Golovleva, L. | | Deposit date: | 2010-07-29 | | Release date: | 2011-08-17 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | X-ray structures of 4-chlorocatechol 1,2-dioxygenase adducts with substituted catechols: new perspectives in the molecular basis of intradiol ring cleaving dioxygenases specificity.
J. Struct. Biol., 181, 2013
|
|
