8PIE
 
 | | Crystal structure of the human nucleoside diphosphate kinase B domain in complex with the product AT-8500 formed by catalysis of compound AT-9010 | | Descriptor: | GLYCEROL, Nucleoside diphosphate kinase B, [(2~{R},3~{R},4~{R},5~{R})-5-(2-azanyl-6-oxidanylidene-1~{H}-purin-9-yl)-4-fluoranyl-4-methyl-3-oxidanyl-oxolan-2-yl]methyl phosphono hydrogen phosphate | | Authors: | Feracci, M, Chazot, A, Ferron, F, Alvarez, K, Canard, B. | | Deposit date: | 2023-06-21 | | Release date: | 2024-07-31 | | Last modified: | 2024-09-04 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The activation cascade of the broad-spectrum antiviral bemnifosbuvir characterized at atomic resolution.
Plos Biol., 22, 2024
|
|
7PLR
 
 | | Crystal structure of the N-terminal endonuclease domain of La Crosse virus L-protein bound to compound Baloxavir | | Descriptor: | Baloxavir acid, FORMIC ACID, GLYCEROL, ... | | Authors: | Feracci, M, Hernandez, S, Vincentelli, R, Ferron, F, Reguera, J, Canard, B, Alvarez, K. | | Deposit date: | 2021-09-01 | | Release date: | 2022-09-14 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.64 Å) | | Cite: | Biophysical and structural study of La Crosse virus endonuclease inhibition for the development of new antiviral options.
Iucrj, 11, 2024
|
|
7OA4
 
 | | Crystal structure of the N-terminal endonuclease domain of La Crosse virus L-protein bound to compound L-742,001 | | Descriptor: | (2Z)-4-[1-benzyl-4-(4-chlorobenzyl)piperidin-4-yl]-2-hydroxy-4-oxobut-2-enoic acid, FORMIC ACID, MANGANESE (II) ION, ... | | Authors: | Feracci, M, Hernandez, S, Vincentelli, R, Ferron, F, Reguera, J, Canard, B, Alvarez, K. | | Deposit date: | 2021-04-19 | | Release date: | 2022-08-03 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Biophysical and structural study of La Crosse virus endonuclease inhibition for the development of new antiviral options.
Iucrj, 11, 2024
|
|
6R9H
 
 | | Crystal structure of the PDZ tandem of syntenin in complex with fragment C58 | | Descriptor: | (2~{S})-2-[2-(4-chlorophenyl)sulfanylethanoylamino]-3-methyl-butanoic acid, ACETATE ION, PHOSPHATE ION, ... | | Authors: | Feracci, M, Barral, K. | | Deposit date: | 2019-04-03 | | Release date: | 2021-02-03 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Pharmacological inhibition of syntenin PDZ2 domain impairs breast cancer cell activities and exosome loadifing with syndecan and EpCAM cargo.
J Extracell Vesicles, 10, 2020
|
|
6RLC
 
 | | Crystal structure of the PDZ tandem of syntenin in complex with fragment F13 | | Descriptor: | (2~{S})-2-[3-(4-chlorophenyl)sulfanylpropanoylamino]-3-methyl-butanoic acid, ACETATE ION, Syntenin-1 | | Authors: | Feracci, M, Barral, K. | | Deposit date: | 2019-05-02 | | Release date: | 2021-02-03 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Pharmacological inhibition of syntenin PDZ2 domain impairs breast cancer cell activities and exosome loadifing with syndecan and EpCAM cargo.
J Extracell Vesicles, 10, 2020
|
|
8PYW
 
 | | Crystal structure of the human Nucleoside-diphosphate kinase B domain bound to compound diphosphate form of AT-9052-Sp. | | Descriptor: | GLYCEROL, Nucleoside diphosphate kinase B, [[(2R,3R,4R,5R)-5-(2-azanyl-6-oxidanylidene-1H-purin-9-yl)-4-fluoranyl-4-methyl-3-oxidanyl-oxolan-2-yl]methoxy-sulfanyl-phosphoryl] dihydrogen phosphate | | Authors: | Feracci, M, Chazot, A. | | Deposit date: | 2023-07-26 | | Release date: | 2023-12-27 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.553 Å) | | Cite: | An exonuclease-resistant chain-terminating nucleotide analogue targeting the SARS-CoV-2 replicase complex.
Nucleic Acids Res., 52, 2024
|
|
8AAK
 
 | | Crystal structure of the PDZ tandem of syntenin in complex with compound 29 | | Descriptor: | (2~{S})-2-[[(2~{S})-2-(3-oxidanylidene-1~{H}-isoindol-2-yl)-3-phenyl-propanoyl]amino]propanoic acid, GLYCEROL, Syntenin-1 | | Authors: | Feracci, M, Barral, K. | | Deposit date: | 2022-07-01 | | Release date: | 2023-04-26 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.554 Å) | | Cite: | Discovery of a PDZ Domain Inhibitor Targeting the Syndecan/Syntenin Protein-Protein Interaction: A Semi-Automated "Hit Identification-to-Optimization" Approach.
J.Med.Chem., 66, 2023
|
|
8AAP
 
 | | Crystal structure of the PDZ tandem of syntenin in complex with compound SYNTi | | Descriptor: | (2S)-2-[[(2S)-2-(6-bromanyl-3-oxidanylidene-1H-isoindol-2-yl)-3-[4-(5-ethanoyl-2-fluoranyl-phenyl)phenyl]propanoyl]amino]propanoic acid, Syntenin-1 | | Authors: | Feracci, M, Barral, K. | | Deposit date: | 2022-07-01 | | Release date: | 2023-04-26 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.174 Å) | | Cite: | Discovery of a PDZ Domain Inhibitor Targeting the Syndecan/Syntenin Protein-Protein Interaction: A Semi-Automated "Hit Identification-to-Optimization" Approach.
J.Med.Chem., 66, 2023
|
|
8AAO
 
 | | Crystal structure of the PDZ tandem of syntenin in complex with compound 95 | | Descriptor: | (2~{S})-2-[[(2~{S})-3-[4-(5-ethanoyl-2-fluoranyl-phenyl)phenyl]-2-(3-oxidanylidene-1~{H}-isoindol-2-yl)propanoyl]amino]propanoic acid, Syntenin-1 | | Authors: | Feracci, M, Barral, K. | | Deposit date: | 2022-07-01 | | Release date: | 2023-04-26 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.469 Å) | | Cite: | Discovery of a PDZ Domain Inhibitor Targeting the Syndecan/Syntenin Protein-Protein Interaction: A Semi-Automated "Hit Identification-to-Optimization" Approach.
J.Med.Chem., 66, 2023
|
|
8AAI
 
 | | Crystal structure of the PDZ tandem of syntenin in complex with fragment E5 | | Descriptor: | (2~{S})-2-[[(2~{S})-3-methyl-2-(3-oxidanylidene-1~{H}-isoindol-2-yl)butanoyl]amino]propanoic acid, Syntenin-1 | | Authors: | Feracci, M, Barral, K. | | Deposit date: | 2022-07-01 | | Release date: | 2023-04-26 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.76 Å) | | Cite: | Discovery of a PDZ Domain Inhibitor Targeting the Syndecan/Syntenin Protein-Protein Interaction: A Semi-Automated "Hit Identification-to-Optimization" Approach.
J.Med.Chem., 66, 2023
|
|
2LKQ
 
 | | NMR structure of the lambda 5 22-45 peptide | | Descriptor: | Immunoglobulin lambda-like polypeptide 1 | | Authors: | Elantak, L, Espeli, M, Boned, A, Bornet, O, Breton, C, Feracci, M, Roche, P, Guerlesquin, F, Schiff, C. | | Deposit date: | 2011-10-19 | | Release date: | 2012-10-24 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural Basis for Galectin-1-dependent Pre-B Cell Receptor (Pre-BCR) Activation.
J.Biol.Chem., 287, 2012
|
|
7ED5
 
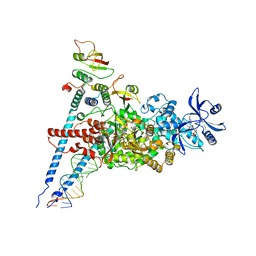 | | A dual mechanism of action of AT-527 against SARS-CoV-2 polymerase | | Descriptor: | MAGNESIUM ION, Non-structural protein 7, Non-structural protein 8, ... | | Authors: | Shannon, A, Fattorini, V, Sama, B, Selisko, B, Feracci, M, Falcou, C, Gauffre, P, El Kazzi, P, Delpal, A, Decroly, E, Alvarez, K, Eydoux, C, Guillemot, J.-C, Moussa, A, Good, S, Colla, P, Lin, K, Sommadossi, J.-P, Zhu, Y.X, Yan, X.D, Shi, H, Ferron, F, Canard, B. | | Deposit date: | 2021-03-15 | | Release date: | 2022-02-16 | | Last modified: | 2024-04-10 | | Method: | ELECTRON MICROSCOPY (2.98 Å) | | Cite: | A dual mechanism of action of AT-527 against SARS-CoV-2 polymerase.
Nat Commun, 13, 2022
|
|
5ELR
 
 | |
5ELS
 
 | |
5EL3
 
 | | Structure of the KH domain of T-STAR | | Descriptor: | KH domain-containing, RNA-binding, signal transduction-associated protein 3, ... | | Authors: | Dominguez, C, Feracci, M. | | Deposit date: | 2015-11-04 | | Release date: | 2016-01-13 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Structural basis of RNA recognition and dimerization by the STAR proteins T-STAR and Sam68.
Nat Commun, 7, 2016
|
|
5ELT
 
 | |
5EMO
 
 | |
