5E6Y
 
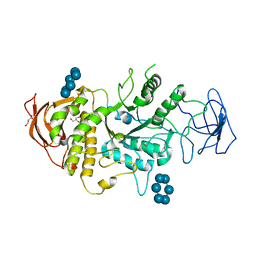 | | Crystal structure of E.Coli branching enzyme in complex with alpha cyclodextrin | | 分子名称: | 1,4-alpha-glucan branching enzyme GlgB, Cyclohexakis-(1-4)-(alpha-D-glucopyranose), GLYCEROL | | 著者 | Feng, L, Nosrati, M, Geiger, J.H. | | 登録日 | 2015-10-11 | | 公開日 | 2015-12-16 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Crystal structures of Escherichia coli branching enzyme in complex with cyclodextrins.
Acta Crystallogr D Struct Biol, 72, 2016
|
|
3ORG
 
 | |
6WDO
 
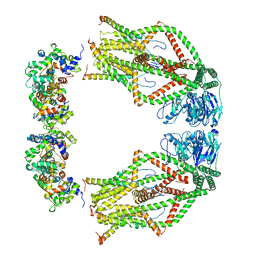 | | Cryo-EM structure of mitochondrial calcium uniporter holocomplex in high Ca2+ | | 分子名称: | CALCIUM ION, Calcium uniporter protein, mitochondrial, ... | | 著者 | Feng, L, Zhang, J, Fan, M. | | 登録日 | 2020-04-01 | | 公開日 | 2020-05-27 | | 最終更新日 | 2024-03-06 | | 実験手法 | ELECTRON MICROSCOPY (3.6 Å) | | 主引用文献 | Structure and mechanism of the mitochondrial Ca2+uniporter holocomplex.
Nature, 582, 2020
|
|
4LQ1
 
 | |
4LPC
 
 | |
6NPK
 
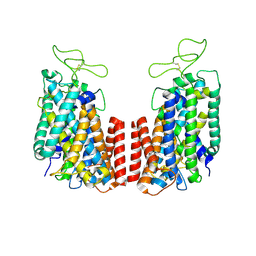 | | Structure of the TM domain | | 分子名称: | Solute carrier family 12 (sodium/potassium/chloride transporter), member 2 | | 著者 | Feng, L, Liao, M.F, Orlando, B, Zhang, J.R. | | 登録日 | 2019-01-17 | | 公開日 | 2019-07-31 | | 最終更新日 | 2019-08-28 | | 実験手法 | ELECTRON MICROSCOPY (3.6 Å) | | 主引用文献 | Structure and mechanism of the cation-chloride cotransporter NKCC1.
Nature, 572, 2019
|
|
6NPJ
 
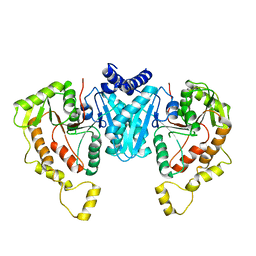 | |
6NPL
 
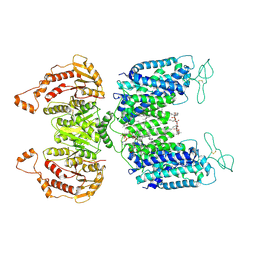 | | Cryo-EM structure of NKCC1 | | 分子名称: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, CHLORIDE ION, POTASSIUM ION, ... | | 著者 | Feng, L, Liao, M.F, Orlando, B, Zhang, J.R. | | 登録日 | 2019-01-17 | | 公開日 | 2019-07-31 | | 最終更新日 | 2019-08-28 | | 実験手法 | ELECTRON MICROSCOPY (2.9 Å) | | 主引用文献 | Structure and mechanism of the cation-chloride cotransporter NKCC1.
Nature, 572, 2019
|
|
6NPH
 
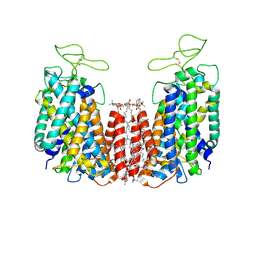 | | Structure of NKCC1 TM domain | | 分子名称: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, CHLORIDE ION, POTASSIUM ION, ... | | 著者 | Feng, L, Liao, M.F, Orlando, B, Zhang, J.R. | | 登録日 | 2019-01-17 | | 公開日 | 2019-07-31 | | 最終更新日 | 2020-01-29 | | 実験手法 | ELECTRON MICROSCOPY (2.9 Å) | | 主引用文献 | Structure and mechanism of the cation-chloride cotransporter NKCC1.
Nature, 572, 2019
|
|
4O0J
 
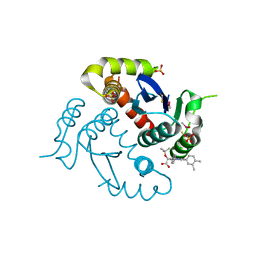 | | HIV-1 Integrase Catalytic Core Domain Complexed with Allosteric Inhibitor (2S)-tert-butoxy[4-(4-chlorophenyl)-6-(3,4-dimethylphenyl)-2,5-dimethylpyridin-3-yl]ethanoic acid | | 分子名称: | (2S)-tert-butoxy[4-(4-chlorophenyl)-6-(3,4-dimethylphenyl)-2,5-dimethylpyridin-3-yl]ethanoic acid, Integrase, SULFATE ION | | 著者 | Feng, L, Kvaratskhelia, M. | | 登録日 | 2013-12-13 | | 公開日 | 2014-07-02 | | 最終更新日 | 2017-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.05 Å) | | 主引用文献 | A New Class of Multimerization Selective Inhibitors of HIV-1 Integrase.
Plos Pathog., 10, 2014
|
|
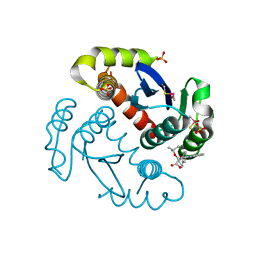
4ID1
 
 | |
4GVM
 
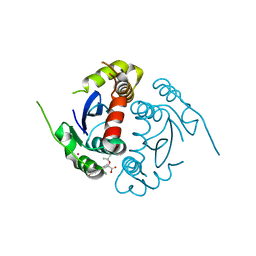 | |
4GW6
 
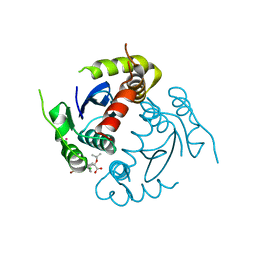 | |
4TSX
 
 | |
4JLH
 
 | |
5JL4
 
 | |
8INJ
 
 | | Crystal structure of UGT74AN3-UDP-DIG | | 分子名称: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, DIGITOXIGENIN, Glycosyltransferase, ... | | 著者 | Feng, L, Wei, H. | | 登録日 | 2023-03-10 | | 公開日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (1.76 Å) | | 主引用文献 | Crystal structure of UGT74AN3-UDP-DIG
To Be Published
|
|
8JJQ
 
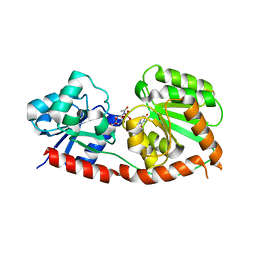 | |
8IN7
 
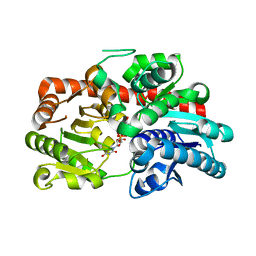 | |
4RZE
 
 | | Crystal Structure Analysis of the NUR77 Ligand Binding Domain, L437W,D594E mutant | | 分子名称: | GLYCEROL, Nuclear receptor subfamily 4 group A member 1 | | 著者 | Fengwei, L, Xuyang, T, Anzhong, L, Li, L, Yuan, L, Hangzi, C, Qiao, W, Tianwei, L. | | 登録日 | 2014-12-21 | | 公開日 | 2015-03-18 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.49 Å) | | 主引用文献 | Impeding the interaction between Nur77 and p38 reduces LPS-induced inflammation.
Nat.Chem.Biol., 11, 2015
|
|
8DYP
 
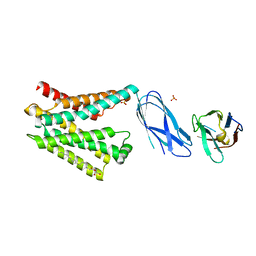 | |
7N98
 
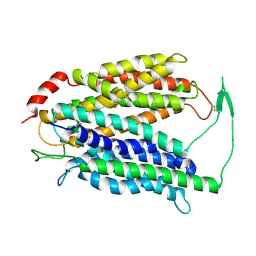 | | Cryo-EM structure of MFSD2A | | 分子名称: | Sodium-dependent lysophosphatidylcholine symporter 1 | | 著者 | Zhang, J, Feng, L. | | 登録日 | 2021-06-17 | | 公開日 | 2021-08-04 | | 最終更新日 | 2021-09-01 | | 実験手法 | ELECTRON MICROSCOPY (3.5 Å) | | 主引用文献 | Structure and mechanism of blood-brain-barrier lipid transporter MFSD2A.
Nature, 596, 2021
|
|
7SL9
 
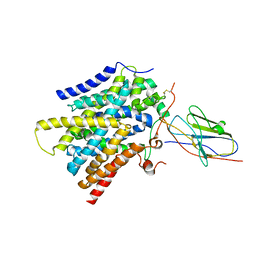 | | CryoEM structure of SMCT1 | | 分子名称: | Sodium-coupled monocarboxylate transporter 1, butanoic acid, nanobody Nb2 | | 著者 | Qu, Q, Han, L, Panova, O, Feng, L, Skiniotis, G. | | 登録日 | 2021-10-23 | | 公開日 | 2021-12-15 | | 最終更新日 | 2022-01-26 | | 実験手法 | ELECTRON MICROSCOPY (3.5 Å) | | 主引用文献 | Structure and mechanism of the SGLT family of glucose transporters.
Nature, 601, 2022
|
|
7SLA
 
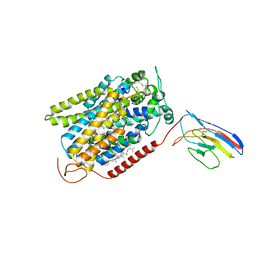 | | CryoEM structure of SGLT1 at 3.15 Angstrom resolution | | 分子名称: | CHOLESTEROL HEMISUCCINATE, Sodium/glucose cotransporter 1, nanobody Nb1 | | 著者 | Qu, Q, Han, L, Panova, O, Feng, L, Skiniotis, G. | | 登録日 | 2021-10-23 | | 公開日 | 2021-12-15 | | 最終更新日 | 2022-01-26 | | 実験手法 | ELECTRON MICROSCOPY (3.15 Å) | | 主引用文献 | Structure and mechanism of the SGLT family of glucose transporters.
Nature, 601, 2022
|
|
7SL8
 
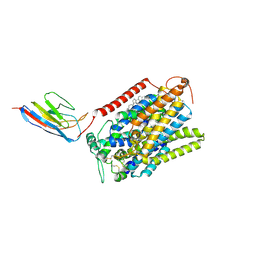 | | CryoEM structure of SGLT1 at 3.4 A resolution | | 分子名称: | CHOLESTEROL, Sodium/glucose cotransporter 1, nanobody Nb1 | | 著者 | Qu, Q, Han, L, Panova, O, Feng, L, Skiniotis, G. | | 登録日 | 2021-10-23 | | 公開日 | 2021-12-15 | | 最終更新日 | 2022-01-26 | | 実験手法 | ELECTRON MICROSCOPY (3.4 Å) | | 主引用文献 | Structure and mechanism of the SGLT family of glucose transporters.
Nature, 601, 2022
|
|
