1B2W
 
 | | COMPARISON OF THE THREE-DIMENSIONAL STRUCTURES OF A HUMANIZED AND A CHIMERIC FAB OF AN ANTI-GAMMA-INTERFERON ANTIBODY | | Descriptor: | PROTEIN (ANTIBODY (HEAVY CHAIN)), PROTEIN (ANTIBODY (LIGHT CHAIN)) | | Authors: | Fan, Z, Shan, L, Goldsteen, B.Z, Guddat, L.W, Thakur, A, Landolfi, N.F, Co, M.S, Vasquez, M, Queen, C, Ramsland, P.A, Edmundson, A.B. | | Deposit date: | 1998-12-01 | | Release date: | 1999-05-06 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Comparison of the three-dimensional structures of a humanized and a chimeric Fab of an anti-gamma-interferon antibody.
J.Mol.Recog., 12, 1999
|
|
1B4J
 
 | | COMPARISON OF THE THREE-DIMENSIONAL STRUCTURES OF A HUMANIZED AND A CHIMERIC FAB OF AN ANTI-GAMMA-INTERFERON ANTIBODY | | Descriptor: | ANTIBODY | | Authors: | Fan, Z, Shan, L, Goldsteen, B.Z, Guddat, L.W, Thakur, A, Landolfi, N.F, Co, M.S, Vasques, M, Queen, C, Ramsland, P.A, Edmundson, A.B. | | Deposit date: | 1998-12-22 | | Release date: | 1999-06-15 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Comparison of the three-dimensional structures of a humanized and a chimeric Fab of an anti-gamma-interferon antibody.
J.Mol.Recog., 12, 1999
|
|
5W7N
 
 | |
6F52
 
 | |
6F4W
 
 | | Crystal structure of H. pylori purine nucleoside phosphorylase in complex with formycin A | | Descriptor: | (1S)-1-(7-amino-1H-pyrazolo[4,3-d]pyrimidin-3-yl)-1,4-anhydro-D-ribitol, Purine nucleoside phosphorylase DeoD-type | | Authors: | Stefanic, Z. | | Deposit date: | 2017-11-30 | | Release date: | 2018-02-14 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.293 Å) | | Cite: | Helicobacter pylori purine nucleoside phosphorylase shows new distribution patterns of open and closed active site conformations and unusual biochemical features.
FEBS J., 285, 2018
|
|
6F5I
 
 | | Crystal structure of H. pylori purine nucleoside phosphorylase | | Descriptor: | METHANOL, Purine nucleoside phosphorylase DeoD-type | | Authors: | Stefanic, Z. | | Deposit date: | 2017-12-01 | | Release date: | 2018-02-14 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.298 Å) | | Cite: | Helicobacter pylori purine nucleoside phosphorylase shows new distribution patterns of open and closed active site conformations and unusual biochemical features.
FEBS J., 285, 2018
|
|
6F5A
 
 | | Crystal structure of H. pylori purine nucleoside phosphorylase | | Descriptor: | Purine nucleoside phosphorylase DeoD-type | | Authors: | Stefanic, Z. | | Deposit date: | 2017-12-01 | | Release date: | 2018-02-14 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.201 Å) | | Cite: | Helicobacter pylori purine nucleoside phosphorylase shows new distribution patterns of open and closed active site conformations and unusual biochemical features.
FEBS J., 285, 2018
|
|
6F4X
 
 | |
6G7X
 
 | |
5USG
 
 | | 5-Se-T2/4-DNA and native RNA hybrid in complex with RNase H catalytic domain D132N mutant | | Descriptor: | DNA (5'-D(*AP*(T5S)P*GP*(T5S)P*CP*G)-3'), MAGNESIUM ION, RNA (5'-R(*UP*CP*GP*AP*CP*A)-3'), ... | | Authors: | Fang, Z, Qin, L, Huang, Z. | | Deposit date: | 2017-02-13 | | Release date: | 2017-03-08 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Synthesis of Pyrimidine Modified Seleno-DNA as a Novel Approach to Antisense Candidate
Chemistryselect, 8, 2023
|
|
5US2
 
 | |
5USA
 
 | | 5-Se-T2-DNA and native RNA hybrid in complex with RNase H catalytic domain D132N mutant | | Descriptor: | DNA (5'-D(*AP*(T5S)P*GP*TP*CP*G)-3'), MAGNESIUM ION, RNA (5'-R(*UP*CP*GP*AP*CP*A)-3'), ... | | Authors: | Fang, Z, Qin, L, Huang, Z. | | Deposit date: | 2017-02-13 | | Release date: | 2017-03-08 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Synthesis of Pyrimidine Modified Seleno-DNA as a Novel Approach to Antisense Candidate
Chemistryselect, 8, 2023
|
|
5USE
 
 | | 5-Se-T4-DNA and native RNA hybrid in complex with RNase H catalytic domain D132N mutant | | Descriptor: | DNA (5'-D(*AP*TP*GP*(T5S)P*CP*G)-3'), MAGNESIUM ION, RNA (5'-R(*UP*CP*GP*AP*CP*A)-3'), ... | | Authors: | Fang, Z, Qin, L, Huang, Z. | | Deposit date: | 2017-02-13 | | Release date: | 2017-03-08 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Synthesis of Pyrimidine Modified Seleno-DNA as a Novel Approach to Antisense Candidate
Chemistryselect, 8, 2023
|
|
5LU0
 
 | | Crystal structure of H. pylori referent strain in complex with PO4 | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, PHOSPHATE ION, ... | | Authors: | Stefanic, Z. | | Deposit date: | 2016-09-07 | | Release date: | 2017-09-20 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Helicobacter pylori purine nucleoside phosphorylase shows new distribution patterns of open and closed active site conformations and unusual biochemical features.
FEBS J., 285, 2018
|
|
5MAL
 
 | |
5MX6
 
 | |
5MX8
 
 | |
5MX4
 
 | |
6PTS
 
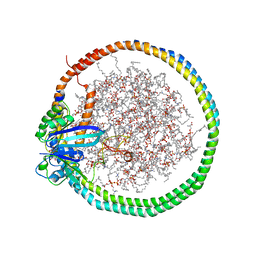 | | NMR data-driven model of KRas-GMPPNP:RBD-CRD complex tethered to a nanodisc (state A) | | Descriptor: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, Apolipoprotein A-I, GTPase KRas, ... | | Authors: | Fang, Z, Lee, K, Gasmi-Seabrook, G, Ikura, M, Marshall, C.B. | | Deposit date: | 2019-07-16 | | Release date: | 2020-05-13 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Multivalent assembly of KRAS with the RAS-binding and cysteine-rich domains of CRAF on the membrane.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6PTW
 
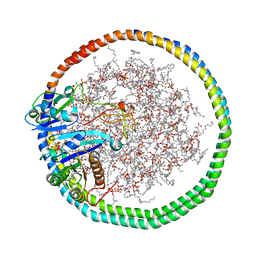 | | NMR data-driven model of KRas-GMPPNP:RBD-CRD complex tethered to a nanodisc (state B) | | Descriptor: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, Apolipoprotein A-I, GTPase KRas, ... | | Authors: | Fang, Z, Lee, K, Gasmi-Seabrook, G, Ikura, M, Marshall, C.B. | | Deposit date: | 2019-07-16 | | Release date: | 2020-05-13 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Multivalent assembly of KRAS with the RAS-binding and cysteine-rich domains of CRAF on the membrane.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
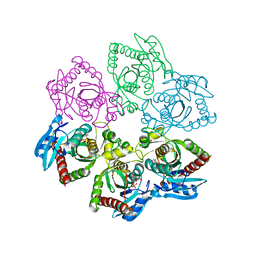
4TS9
 
 | |
4TTJ
 
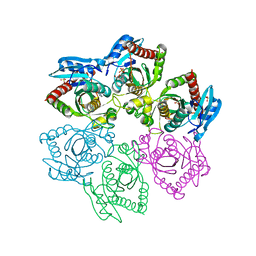 | | Crystal structure of double mutant E. Coli purine nucleoside phosphorylase with 6 FMC molecules | | Descriptor: | (1S)-1-(7-amino-1H-pyrazolo[4,3-d]pyrimidin-3-yl)-1,4-anhydro-D-ribitol, PHOSPHATE ION, Purine nucleoside phosphorylase DeoD-type, ... | | Authors: | Stefanic, Z, Bzowska, A. | | Deposit date: | 2014-06-21 | | Release date: | 2015-07-08 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.874 Å) | | Cite: | Crystallographic snapshots of ligand binding to hexameric purine nucleoside phosphorylase and kinetic studies give insight into the mechanism of catalysis.
Sci Rep, 8, 2018
|
|
4TTA
 
 | | Crystal structure of double mutant E. Coli purine nucleoside phosphorylase with 2 FMC molecules | | Descriptor: | (1S)-1-(7-amino-1H-pyrazolo[4,3-d]pyrimidin-3-yl)-1,4-anhydro-D-ribitol, PHOSPHATE ION, Purine nucleoside phosphorylase DeoD-type, ... | | Authors: | Stefanic, Z, Bzowska, A. | | Deposit date: | 2014-06-20 | | Release date: | 2015-07-08 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystallographic snapshots of ligand binding to hexameric purine nucleoside phosphorylase and kinetic studies give insight into the mechanism of catalysis.
Sci Rep, 8, 2018
|
|
4TS3
 
 | |
4TTI
 
 | | Crystal structure of double mutant E. Coli purine nucleoside phosphorylase with 4 FMC molecules | | Descriptor: | (1S)-1-(7-amino-1H-pyrazolo[4,3-d]pyrimidin-3-yl)-1,4-anhydro-D-ribitol, PHOSPHATE ION, Purine nucleoside phosphorylase DeoD-type, ... | | Authors: | Stefanic, Z, Bzowska, A. | | Deposit date: | 2014-06-21 | | Release date: | 2015-07-08 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Crystallographic snapshots of ligand binding to hexameric purine nucleoside phosphorylase and kinetic studies give insight into the mechanism of catalysis.
Sci Rep, 8, 2018
|
|
