2KBE
 
 | |
6V8Q
 
 | | Structure of an inner membrane protein required for PhoPQ regulated increases in outer membrane cardiolipin | | Descriptor: | (9Z,21R,24R,30R,33R,44Z)-24,27,30-trihydroxy-18,24,30,36-tetraoxo-19,23,25,29,31,35-hexaoxa-24lambda~5~,30lambda~5~-dip hosphatripentaconta-9,44-diene-21,33-diyl (9Z,9'Z)di-octadec-9-enoate, CALCIUM ION, DODECYL-BETA-D-MALTOSIDE, ... | | Authors: | Fan, J, Miller, S. | | Deposit date: | 2019-12-11 | | Release date: | 2020-03-18 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.699585 Å) | | Cite: | Structure of an Inner Membrane Protein Required for PhoPQ-Regulated Increases in Outer Membrane Cardiolipin.
Mbio, 11, 2020
|
|
2KBF
 
 | |
7BUT
 
 | |
8JQR
 
 | | Structure of human TRPV1 in complex with antagonist | | Descriptor: | 4-(7-Hydroxy-2-isopropyl-4-oxoquinazolin-3(4H)-yl)benzonitrile, CHOLESTEROL, Transient receptor potential cation channel subfamily V member 1,PreScission Site,Green fluorescent protein | | Authors: | Fan, J, Lei, X. | | Deposit date: | 2023-06-14 | | Release date: | 2024-08-14 | | Last modified: | 2024-08-21 | | Method: | ELECTRON MICROSCOPY (2.75 Å) | | Cite: | Structural basis of TRPV1 inhibition by SAF312 and cholesterol.
Nat Commun, 15, 2024
|
|
8JU6
 
 | | Structure of human TRPV4 with antagonist GSK279 | | Descriptor: | 1-({(5S,7S)-3-[5-(2-hydroxypropan-2-yl)pyrazin-2-yl]-7-methyl-2-oxo-1-oxa-3-azaspiro[4.5]decan-7-yl}methyl)-1H-benzimidazole-6-carbonitrile, Transient receptor potential cation channel subfamily V member 4,3C-GFP | | Authors: | Fan, J, Lei, X. | | Deposit date: | 2023-06-24 | | Release date: | 2024-05-08 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.45 Å) | | Cite: | Structural Pharmacology of TRPV4 Antagonists.
Adv Sci, 11, 2024
|
|
8JVJ
 
 | | Structure of human TRPV4 with antagonist A2 and RhoA | | Descriptor: | Transforming protein RhoA, Transient receptor potential cation channel subfamily V member 4,3C-GFP, [6-[[4-(2,4-dimethyl-1,3-thiazol-5-yl)-1,3-thiazol-2-yl]amino]pyridin-3-yl]-[(1~{S},5~{R})-3-[5-(trifluoromethyl)pyrimidin-2-yl]-3,8-diazabicyclo[3.2.1]octan-8-yl]methanone | | Authors: | Fan, J, Lei, X. | | Deposit date: | 2023-06-28 | | Release date: | 2024-05-08 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.44 Å) | | Cite: | Structural Pharmacology of TRPV4 Antagonists.
Adv Sci, 11, 2024
|
|
8JU5
 
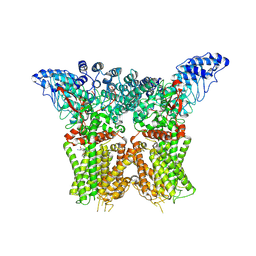 | | Structure of human TRPV4 with antagonist A1 | | Descriptor: | 4-[(3~{S},4~{S})-4-(aminomethyl)-1-(5-chloranylpyridin-2-yl)sulfonyl-4-oxidanyl-pyrrolidin-3-yl]oxy-2-fluoranyl-benzenecarbonitrile, Transient receptor potential cation channel subfamily V member 4,3C-GFP | | Authors: | Fan, J, Lei, X. | | Deposit date: | 2023-06-24 | | Release date: | 2024-05-08 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.74 Å) | | Cite: | Structural Pharmacology of TRPV4 Antagonists.
Adv Sci, 11, 2024
|
|
8JVI
 
 | | Structure of human TRPV4 with antagonist A2 | | Descriptor: | Transient receptor potential cation channel subfamily V member 4,3C-GFP, [6-[[4-(2,4-dimethyl-1,3-thiazol-5-yl)-1,3-thiazol-2-yl]amino]pyridin-3-yl]-[(1~{S},5~{R})-3-[5-(trifluoromethyl)pyrimidin-2-yl]-3,8-diazabicyclo[3.2.1]octan-8-yl]methanone | | Authors: | Fan, J, Lei, X. | | Deposit date: | 2023-06-28 | | Release date: | 2024-05-08 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.21 Å) | | Cite: | Structural Pharmacology of TRPV4 Antagonists.
Adv Sci, 11, 2024
|
|
8WR2
 
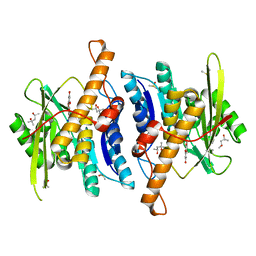 | | Crystal Structure of Human Pyridoxal Kinase with bound Luteolin | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-(3,4-dihydroxyphenyl)-5,7-dihydroxy-4H-chromen-4-one, DIMETHYL SULFOXIDE, ... | | Authors: | Fan, J, Zhu, Y. | | Deposit date: | 2023-10-12 | | Release date: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Discovery and characterization of natural product luteolin as an effective inhibitor of human pyridoxal kinase.
Bioorg.Chem., 143, 2024
|
|
7XJ2
 
 | | Structure of human TRPV3_G573S in complex with Trpvicin in C4 symmetry | | Descriptor: | Fusion protein of Transient receptor potential cation channel subfamily V member 3 and 3C-GFP, N-[5-[2-(2-cyanopropan-2-yl)pyridin-4-yl]-4-(trifluoromethyl)-1,3-thiazol-2-yl]-4,6-dimethoxy-pyrimidine-5-carboxamide | | Authors: | Fan, J, Yue, Z, Jiang, D, Lei, X. | | Deposit date: | 2022-04-14 | | Release date: | 2022-11-09 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.64 Å) | | Cite: | Structural basis of TRPV3 inhibition by an antagonist.
Nat.Chem.Biol., 19, 2023
|
|
7XJ1
 
 | | Structure of human TRPV3_G573S in complex with Trpvicin in C2 symmetry | | Descriptor: | Fusion protein of Transient receptor potential cation channel subfamily V member 3 and 3C-GFP, N-[5-[2-(2-cyanopropan-2-yl)pyridin-4-yl]-4-(trifluoromethyl)-1,3-thiazol-2-yl]-4,6-dimethoxy-pyrimidine-5-carboxamide, [(2~{R})-1-[2-azanylethoxy(oxidanyl)phosphoryl]oxy-3-hexadecanoyloxy-propan-2-yl] (~{Z})-octadec-9-enoate | | Authors: | Fan, J, Yue, Z, Jiang, D, Lei, X. | | Deposit date: | 2022-04-14 | | Release date: | 2022-11-09 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (2.93 Å) | | Cite: | Structural basis of TRPV3 inhibition by an antagonist.
Nat.Chem.Biol., 19, 2023
|
|
7XJ3
 
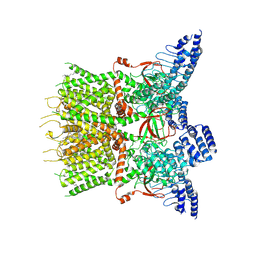 | | Structure of human TRPV3 | | Descriptor: | [(2~{R})-1-[2-azanylethoxy(oxidanyl)phosphoryl]oxy-3-hexadecanoyloxy-propan-2-yl] (~{Z})-octadec-9-enoate, fusion of transient receptor potential cation channel subfamily V member 3 and 3C-GFP | | Authors: | Fan, J, Yue, Z, Jiang, D, Lei, X. | | Deposit date: | 2022-04-14 | | Release date: | 2022-11-09 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.54 Å) | | Cite: | Structural basis of TRPV3 inhibition by an antagonist.
Nat.Chem.Biol., 19, 2023
|
|
7XJ0
 
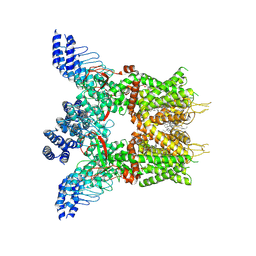 | | Structure of human TRPV3 in complex with Trpvicin | | Descriptor: | Fusion protein of Transient receptor potential cation channel subfamily V member 3 and 3C-GFP, N-[5-[2-(2-cyanopropan-2-yl)pyridin-4-yl]-4-(trifluoromethyl)-1,3-thiazol-2-yl]-4,6-dimethoxy-pyrimidine-5-carboxamide, [(2~{R})-1-[2-azanylethoxy(oxidanyl)phosphoryl]oxy-3-hexadecanoyloxy-propan-2-yl] (~{Z})-octadec-9-enoate | | Authors: | Fan, J, Yue, Z, Jiang, D, Lei, X. | | Deposit date: | 2022-04-14 | | Release date: | 2022-11-09 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.53 Å) | | Cite: | Structural basis of TRPV3 inhibition by an antagonist.
Nat.Chem.Biol., 19, 2023
|
|
5YDX
 
 | |
5YDY
 
 | |
1F96
 
 | | SOLUTION STRUCTURE OF DYNEIN LIGHT CHAIN 8 (DLC8) AND NNOS PEPTIDE COMPLEX | | Descriptor: | DYNEIN LIGHT CHAIN 8, PROTEIN (NNOS, NEURONAL NITRIC OXIDE SYNTHASE) | | Authors: | Fan, J.S, Zhang, Q, Tochio, H, Li, M, Zhang, M. | | Deposit date: | 2000-07-07 | | Release date: | 2001-02-28 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structural basis of diverse sequence-dependent target recognition by the 8 kDa dynein light chain.
J.Mol.Biol., 306, 2001
|
|
6LAG
 
 | | Solution structure of SPA-2 SHD | | Descriptor: | Spa2-like protein | | Authors: | Fan, J.S, Wong, J.Y, Zheng, P, Yang, D, Jedd, G. | | Deposit date: | 2019-11-12 | | Release date: | 2020-04-29 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Spitzenkorper assembly mechanisms reveal conserved features of fungal and metazoan polarity scaffolds.
Nat Commun, 11, 2020
|
|
6KLM
 
 | | NMR solution structure of Roseltide rT7 | | Descriptor: | Roseltide rT7 | | Authors: | Fan, J.S, Kam, A, Loo, S, Yang, D, Tam, P.J. | | Deposit date: | 2019-07-30 | | Release date: | 2019-11-20 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | Roseltide rT7 is a disulfide-rich, anionic, and cell-penetrating peptide that inhibits proteasomal degradation.
J.Biol.Chem., 294, 2019
|
|
5H3N
 
 | |
5H3M
 
 | |
8X94
 
 | | Structure of human TRPV1 in complex with antagonist --protein purified without CHS | | Descriptor: | 4-(7-Hydroxy-2-isopropyl-4-oxoquinazolin-3(4H)-yl)benzonitrile, CHOLESTEROL, Transient receptor potential cation channel subfamily V member 1,Green fluorescent protein | | Authors: | Fan, J, Lei, X. | | Deposit date: | 2023-11-29 | | Release date: | 2024-08-14 | | Last modified: | 2024-08-21 | | Method: | ELECTRON MICROSCOPY (2.55 Å) | | Cite: | Structural basis of TRPV1 inhibition by SAF312 and cholesterol.
Nat Commun, 15, 2024
|
|
2M6Z
 
 | |
6LMR
 
 | | Solution structure of cold shock domain and ssDNA complex | | Descriptor: | DNA (5'-D(P*AP*AP*CP*AP*CP*CP*T)-3'), Y-box-binding protein 1 | | Authors: | Fan, J, Yang, D. | | Deposit date: | 2019-12-26 | | Release date: | 2020-07-22 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural basis of DNA binding to human YB-1 cold shock domain regulated by phosphorylation.
Nucleic Acids Res., 48, 2020
|
|
2H35
 
 | | Solution structure of Human normal adult hemoglobin | | Descriptor: | Hemoglobin alpha subunit, Hemoglobin beta subunit, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Fan, J.S, Yang, D. | | Deposit date: | 2006-05-22 | | Release date: | 2006-11-14 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | A new strategy for structure determination of large proteins in solution without deuteration
Nat.Methods, 3, 2006
|
|
