6NWD
 
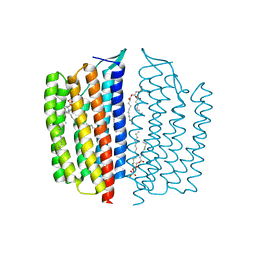 | | X-ray Crystallographic structure of Gloeobacter rhodopsin | | Descriptor: | 1,2-DIMYRISTOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, DECANE, DODECANE, ... | | Authors: | Ernst, O.P, Morizumi, T, Ou, W.L. | | Deposit date: | 2019-02-06 | | Release date: | 2019-08-14 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | X-ray Crystallographic Structure and Oligomerization of Gloeobacter Rhodopsin.
Sci Rep, 9, 2019
|
|
6OFJ
 
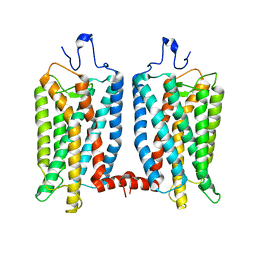 | |
9CDC
 
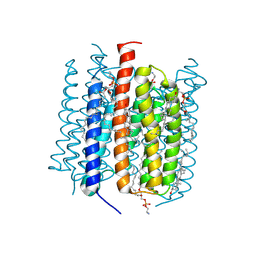 | | Kalium channelrhodopsin 1 C110A mutant from Hyphochytrium catenoides, Dark State | | Descriptor: | 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, CHOLESTEROL, Kalium channelrhodopsin 1 C110A, ... | | Authors: | Morizumi, T, Kim, K, Ernst, O.P. | | Deposit date: | 2024-06-24 | | Release date: | 2025-02-19 | | Method: | ELECTRON MICROSCOPY (3.08 Å) | | Cite: | Structural insights into light-gating of potassium-selective channelrhodopsin.
Nat Commun, 16, 2025
|
|
9CDE
 
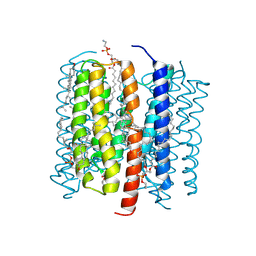 | | Kalium channelrhodopsin 1 C110A mutant from Hyphochytrium catenoides, Continuous Illumination State | | Descriptor: | 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, CHOLESTEROL, Kalium Channelrhodopsin 1 C110A - Continuous Illumination State, ... | | Authors: | Morizumi, T, Kim, K, Ernst, O.P. | | Deposit date: | 2024-06-24 | | Release date: | 2025-02-19 | | Method: | ELECTRON MICROSCOPY (3.44 Å) | | Cite: | Structural insights into light-gating of potassium-selective channelrhodopsin.
Nat Commun, 16, 2025
|
|
9CDD
 
 | | Kalium channelrhodopsin 1 C110A mutant from Hyphochytrium catenoides, Laser-Flash-Illuminated | | Descriptor: | 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, CHOLESTEROL, Kalium Channelrhodopsin 1, ... | | Authors: | Morizumi, T, Kim, K, Ernst, O.P. | | Deposit date: | 2024-06-24 | | Release date: | 2025-02-19 | | Method: | ELECTRON MICROSCOPY (3.05 Å) | | Cite: | Structural insights into light-gating of potassium-selective channelrhodopsin.
Nat Commun, 16, 2025
|
|
5YQZ
 
 | | Structure of the glucagon receptor in complex with a glucagon analogue | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Glucagon analogue, ... | | Authors: | Zhang, H, Qiao, A, Yang, L, VAN EPS, N, Frederiksen, K, Yang, D, Dai, A, Cai, X, Zhang, H, Yi, C, Can, C, He, L, Yang, H, Lau, J, Ernst, O, Hanson, M, Stevens, R, Wang, M, Seedtz-Runge, S, Jiang, H, Zhao, Q, Wu, B. | | Deposit date: | 2017-11-08 | | Release date: | 2018-01-17 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure of the glucagon receptor in complex with a glucagon analogue.
Nature, 553, 2018
|
|
7U55
 
 | | Crystal structure of Thermoplasmatales archaeon heliorhodopsin at pH 4.5 | | Descriptor: | CHLORIDE ION, DODECANE, Heliorhodopsin, ... | | Authors: | Besaw, J.E, De Guzman, P, Miller, R.J.D, Ernst, O.P. | | Deposit date: | 2022-03-01 | | Release date: | 2022-09-07 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Low pH structure of heliorhodopsin reveals chloride binding site and intramolecular signaling pathway.
Sci Rep, 12, 2022
|
|
8GI9
 
 | | Cation channelrhodopsin from Hyphochytrium catenoides (HcCCR) embedded in peptidisc | | Descriptor: | 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, CHOLESTEROL, Cation Channelrhodopsin, ... | | Authors: | Morizumi, T, Kim, K, Li, H, Spudich, J.L, Ernst, O.P. | | Deposit date: | 2023-03-13 | | Release date: | 2023-07-26 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (2.84 Å) | | Cite: | Structures of channelrhodopsin paralogs in peptidiscs explain their contrasting K + and Na + selectivities.
Nat Commun, 14, 2023
|
|
8GI8
 
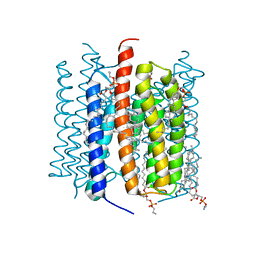 | | Kalium channelrhodopsin 1 from Hyphochytrium catenoides (HcKCR1) embedded in peptidisc | | Descriptor: | 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, CHOLESTEROL, Kalium Channelrhodopsin 1, ... | | Authors: | Morizumi, T, Kim, K, Li, H, Spudich, J.L, Ernst, O.P. | | Deposit date: | 2023-03-13 | | Release date: | 2023-07-26 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (2.88 Å) | | Cite: | Structures of channelrhodopsin paralogs in peptidiscs explain their contrasting K + and Na + selectivities.
Nat Commun, 14, 2023
|
|
8EL2
 
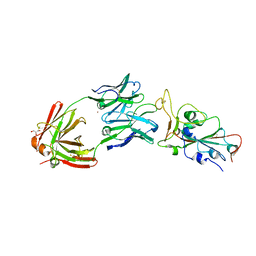 | | SARS-CoV-2 RBD bound to neutralizing antibody Fab ICO-hu23 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Fab ICO-hu23 Heavy Chain, Fab ICO-hu23 Light Chain, ... | | Authors: | Besaw, J.E, Kuo, A, Morizumi, T, Ernst, O.P. | | Deposit date: | 2022-09-22 | | Release date: | 2023-07-19 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.89 Å) | | Cite: | Broadly neutralizing humanized SARS-CoV-2 antibody binds to a conserved epitope on Spike and provides antiviral protection through inhalation-based delivery in non-human primates.
Plos Pathog., 19, 2023
|
|
8ELJ
 
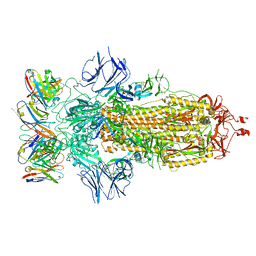 | | SARS-CoV-2 spike glycoprotein in complex with the ICO-hu23 neutralizing antibody Fab fragment | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ICO-hu23 antibody Fab heavy chain, ... | | Authors: | Yee, A.W, Morizumi, T, Kim, K, Kuo, A, Ernst, O.P. | | Deposit date: | 2022-09-24 | | Release date: | 2023-07-19 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Broadly neutralizing humanized SARS-CoV-2 antibody binds to a conserved epitope on Spike and provides antiviral protection through inhalation-based delivery in non-human primates.
Plos Pathog., 19, 2023
|
|
6BK9
 
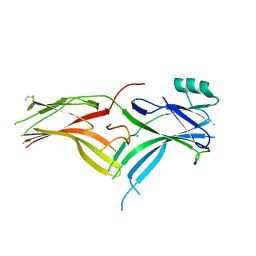 | | Crystal Structure of Squid Arrestin | | Descriptor: | CHLORIDE ION, Visual arrestin | | Authors: | Eger, B.T, Bandyopadhyay, A, Yedidi, R.S, Ernst, O.P. | | Deposit date: | 2017-11-08 | | Release date: | 2018-09-19 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.00005579 Å) | | Cite: | A Novel Polar Core and Weakly Fixed C-Tail in Squid Arrestin Provide New Insight into Interaction with Rhodopsin.
J. Mol. Biol., 430, 2018
|
|
4J4Q
 
 | | Crystal structure of active conformation of GPCR opsin stabilized by octylglucoside | | Descriptor: | ACETATE ION, Guanine nucleotide-binding protein G(t) subunit alpha-1, PALMITIC ACID, ... | | Authors: | Park, J.H, Morizumi, T, Li, Y, Hong, J.E, Pai, E.F, Hofmann, K.P, Choe, H.W, Ernst, O.P. | | Deposit date: | 2013-02-07 | | Release date: | 2013-10-30 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Opsin, a structural model for olfactory receptors?
Angew.Chem.Int.Ed.Engl., 52, 2013
|
|
6V51
 
 | | Spin-labeled T4 Lysozyme (9/131FnbY)-(4-Amino-TEMPO) | | Descriptor: | 4-amino-2,2,6,6-tetramethylpiperidin-1-ol, Endolysin | | Authors: | Liu, J, Morizumi, T, Ou, W.L, Wang, L, Ernst, O.P. | | Deposit date: | 2019-12-02 | | Release date: | 2020-10-07 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Genetically Encoded Quinone Methides Enabling Rapid, Site-Specific, and Photocontrolled Protein Modification with Amine Reagents.
J.Am.Chem.Soc., 142, 2020
|
|
6WP8
 
 | | Proton-pumping mutant of Mastigocladopsis repens rhodopsin chloride pump | | Descriptor: | Proton-pumping rhodopsin chloride pump, RETINAL, octyl beta-D-glucopyranoside | | Authors: | Besaw, J.E, Ernst, O.P, Ou, W, Morizumi, T. | | Deposit date: | 2020-04-26 | | Release date: | 2020-07-29 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The crystal structures of a chloride-pumping microbial rhodopsin and its proton-pumping mutant illuminate proton transfer determinants.
J.Biol.Chem., 295, 2020
|
|
6XL3
 
 | | Mastigocladopsis repens rhodopsin chloride pump | | Descriptor: | CHLORIDE ION, DECANE, Mastigocladopsis repens rhodopsin chloride pump, ... | | Authors: | Besaw, J.E, Ernst, O.P, Ou, W, Morizumi, T. | | Deposit date: | 2020-06-28 | | Release date: | 2020-07-29 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | The crystal structures of a chloride-pumping microbial rhodopsin and its proton-pumping mutant illuminate proton transfer determinants.
J.Biol.Chem., 295, 2020
|
|
7F1X
 
 | | X-ray crystal structure of visual arrestin complexed with inositol 1,4,5-triphosphate | | Descriptor: | 1,2-ETHANEDIOL, D-MYO-INOSITOL-1,4,5-TRIPHOSPHATE, PENTANEDIAL, ... | | Authors: | Jang, K, Kang, M, Eger, B.T, Choe, H.W, Ernst, O.P, Kim, Y.J. | | Deposit date: | 2021-06-10 | | Release date: | 2021-10-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural evidence for visual arrestin priming via complexation of phosphoinositols.
Structure, 30, 2022
|
|
7F1W
 
 | | X-ray crystal structure of visual arrestin complexed with inositol hexaphosphate | | Descriptor: | INOSITOL HEXAKISPHOSPHATE, S-arrestin | | Authors: | Kang, M, Jang, K, Eger, B.T, Ernst, O.P, Choe, H.W, Kim, Y.J. | | Deposit date: | 2021-06-10 | | Release date: | 2021-10-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.097 Å) | | Cite: | Structural evidence for visual arrestin priming via complexation of phosphoinositols.
Structure, 30, 2022
|
|
3PXO
 
 | | Crystal structure of Metarhodopsin II | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, PALMITIC ACID, RETINAL, ... | | Authors: | Choe, H.-W, Kim, Y.J, Park, J.H, Morizumi, T, Pai, E.F, Krauss, N, Hofmann, K.P, Scheerer, P, Ernst, O.P. | | Deposit date: | 2010-12-10 | | Release date: | 2011-03-09 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of metarhodopsin II.
Nature, 471, 2011
|
|
3PQR
 
 | | Crystal structure of Metarhodopsin II in complex with a C-terminal peptide derived from the Galpha subunit of transducin | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, Guanine nucleotide-binding protein G(t) subunit alpha-1, ... | | Authors: | Choe, H.-W, Kim, Y.J, Park, J.H, Morizumi, T, Pai, E.F, Krauss, N, Hofmann, K.P, Scheerer, P, Ernst, O.P. | | Deposit date: | 2010-11-26 | | Release date: | 2011-03-09 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Crystal structure of metarhodopsin II.
Nature, 471, 2011
|
|
3CAP
 
 | | Crystal Structure of Native Opsin: the G Protein-Coupled Receptor Rhodopsin in its Ligand-free State | | Descriptor: | 2-O-octyl-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, PALMITIC ACID, ... | | Authors: | Park, J.H, Scheerer, P, Hofmann, K.P, Choe, H.-W, Ernst, O.P. | | Deposit date: | 2008-02-20 | | Release date: | 2008-06-24 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure of the ligand-free G-protein-coupled receptor opsin
Nature, 454, 2008
|
|
3DQB
 
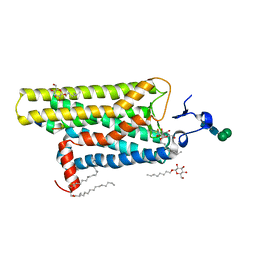 | | Crystal structure of the active G-protein-coupled receptor opsin in complex with a C-terminal peptide derived from the Galpha subunit of transducin | | Descriptor: | 11meric peptide form Guanine nucleotide-binding protein G(t) subunit alpha-1, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, PALMITIC ACID, ... | | Authors: | Scheerer, P, Park, J.H, Hildebrand, P.W, Kim, Y.J, Krauss, N, Choe, H.-W, Hofmann, K.P, Ernst, O.P. | | Deposit date: | 2008-07-09 | | Release date: | 2008-09-23 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystal structure of opsin in its G-protein-interacting conformation
Nature, 455, 2008
|
|
7JJO
 
 | | Structural Basis of the Activation of Heterotrimeric Gs-protein by Isoproterenol-bound Beta1-Adrenergic Receptor | | Descriptor: | Beta1-Adrenergic Receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Su, M, Zhu, L, Zhang, Y, Paknejad, N, Dey, R, Huang, J, Lee, M.Y, Williams, D, Jordan, K.D, Eng, E.T, Ernst, O.P, Meyerson, J.R, Hite, R.K, Walz, T, Liu, W, Huang, X.Y. | | Deposit date: | 2020-07-27 | | Release date: | 2020-09-02 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Structural Basis of the Activation of Heterotrimeric Gs-Protein by Isoproterenol-Bound beta 1 -Adrenergic Receptor.
Mol.Cell, 80, 2020
|
|
6NWE
 
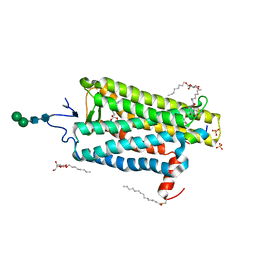 | |
5W0P
 
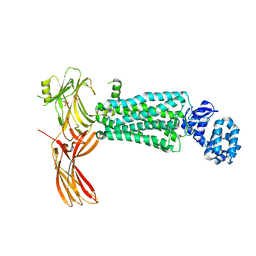 | | Crystal structure of rhodopsin bound to visual arrestin determined by X-ray free electron laser | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Endolysin,Rhodopsin,S-arrestin | | Authors: | Zhou, X.E, He, Y, de Waal, P.W, Gao, X, Kang, Y, Van Eps, N, Yin, Y, Pal, K, Goswami, D, White, T.A, Barty, A, Latorraca, N.R, Chapman, H.N, Hubbell, W.L, Dror, R.O, Stevens, R.C, Cherezov, V, Gurevich, V.V, Griffin, P.R, Ernst, O.P, Melcher, K, Xu, H.E. | | Deposit date: | 2017-05-31 | | Release date: | 2017-08-09 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.013 Å) | | Cite: | Identification of Phosphorylation Codes for Arrestin Recruitment by G Protein-Coupled Receptors.
Cell, 170, 2017
|
|
