1S1F
 
 | | Crystal Structure of Streptomyces Coelicolor A3(2) CYP158A2 from antibiotic biosynthetic pathways | | Descriptor: | 4-PHENYL-1H-IMIDAZOLE, GLYCEROL, MALONIC ACID, ... | | Authors: | Zhao, B, Lamb, D.C, Lei, L, Sundaramoorthy, M, Podust, L.M, Waterman, M.R. | | Deposit date: | 2004-01-06 | | Release date: | 2005-01-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Binding of Two Flaviolin Substrate Molecules, Oxidative Coupling, and Crystal Structure of Streptomyces coelicolor A3(2) Cytochrome P450 158A2
J.Biol.Chem., 280, 2005
|
|
1S6W
 
 | | Solution Structure of hybrid white striped bass hepcidin | | Descriptor: | Hepcidin | | Authors: | Babon, J.J, Singh, S, Pennington, M.W, Norton, R.S, Westerman, M.E. | | Deposit date: | 2004-01-28 | | Release date: | 2004-12-14 | | Last modified: | 2024-10-09 | | Method: | SOLUTION NMR | | Cite: | Bass hepcidin synthesis, solution structure, antimicrobial activities and synergism, and in vivo hepatic response to bacterial infections.
J.Biol.Chem., 280, 2005
|
|
4B7P
 
 | | Structure of HSP90 with NMS-E973 inhibitor bound | | Descriptor: | 5-[2,4-dihydroxy-6-(4-nitrophenoxy)phenyl]-N-(1-methylpiperidin-4-yl)-1,2-oxazole-3-carboxamide, HEAT SHOCK PROTEIN HSP 90-ALPHA | | Authors: | Fogliatto, G, Gianellini, L, Brasca, M.G, Casale, E, Ballinari, D, Ciomei, M, Degrassi, A, De Ponti, A, Germani, M, Guanci, M, Paolucci, M, Polucci, P, Russo, M, Sola, F, Valsasina, B, Visco, C, Zuccotto, F, Donati, D, Felder, E, Galvani, A, Pesenti, E, Mantegani, S, Isacchi, A. | | Deposit date: | 2012-08-21 | | Release date: | 2013-05-29 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Nms-E973, a Novel Synthetic Inhibitor of Hsp90 with Activity in Models of Drug Resistance to Targeted Agents, Including Intracranial Metastases.
Clin.Cancer Res., 19, 2013
|
|
3V4E
 
 | | Crystal Structure of the galactoside O-acetyltransferase in complex with CoA | | Descriptor: | COENZYME A, DI(HYDROXYETHYL)ETHER, Galactoside O-acetyltransferase, ... | | Authors: | Knapik, A.A, Shumilin, I.A, Luo, H.-B, Chruszcz, M, Zimmerman, M.D, Cymborowski, M, Anderson, W.F, Minor, W, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-12-14 | | Release date: | 2012-01-11 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Biophysical analysis of the putative acetyltransferase SACOL2570 from methicillin-resistant Staphylococcus aureus.
J.Struct.Funct.Genom., 14, 2013
|
|
2BON
 
 | | Structure of an Escherichia coli lipid kinase (YegS) | | Descriptor: | LIPID KINASE, MAGNESIUM ION | | Authors: | Bakali, H.M, Johnson, K.A, Hallberg, B.M, Herman, M.D, Nordlund, P. | | Deposit date: | 2005-04-12 | | Release date: | 2006-10-18 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure of Yegs, a Homologue to the Mammalian Diacylglycerol Kinases, Reveals a Novel Regulatory Metal Binding Site.
J.Biol.Chem., 282, 2007
|
|
5AHS
 
 | | 3-Sulfinopropionyl-Coenzyme A (3SP-CoA) desulfinase from Advenella mimgardefordensis DPN7T: holo crystal structure with the substrate analog succinyl-CoA | | Descriptor: | ACYL-COA DEHYDROGENASE, COENZYME A, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Cianci, M, Schuermann, M, Meijers, R, Schneider, T.R, Steinbuechel, A. | | Deposit date: | 2015-02-06 | | Release date: | 2015-06-03 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | 3-Sulfinopropionyl-Coenzyme a (3Sp-Coa) Desulfinase from Advenella Mimigardefordensis Dpn7(T): Crystal Structure and Function of a Desulfinase with an Acyl-Coa Dehydrogenase Fold.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
2CIB
 
 | | High throughput screening and x-ray crystallography assisted evaluation of small molecule scaffolds for CYP51 inhibitors | | Descriptor: | (2S)-2-[(2,1,3-BENZOTHIADIAZOL-4-YLSULFONYL)AMINO]-2-PHENYL-N-PYRIDIN-4-YLACETAMIDE, CYTOCHROME P450 51, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Podust, L.M, Kim, Y, Yermalitskaya, L.V, Von Kries, J.P, Waterman, M.R. | | Deposit date: | 2006-03-17 | | Release date: | 2007-07-03 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Small Molecule Scaffolds for Cyp51 Inhibitors Identified by High Throughput Screening and Defined by X-Ray Crystallography
Antimicrob.Agents Chemother., 51, 2007
|
|
5AF7
 
 | | 3-Sulfinopropionyl-coenzyme A (3SP-CoA) desulfinase from Advenella mimigardefordensis DPN7T: crystal structure and function of a desulfinase with an acyl-CoA dehydrogenase fold. Native crystal structure | | Descriptor: | ACYL-COA DEHYDROGENASE, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL | | Authors: | Cianci, M, Schuermann, M, Meijers, R, Schneider, T.R, Steinbuechel, A. | | Deposit date: | 2015-01-20 | | Release date: | 2015-06-03 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | 3-Sulfinopropionyl-Coenzyme a (3Sp-Coa) Desulfinase from Advenella Mimigardefordensis Dpn7(T): Crystal Structure and Function of a Desulfinase with an Acyl-Coa Dehydrogenase Fold.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
1SSZ
 
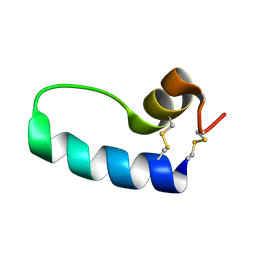 | | Conformational Mapping of Mini-B: An N-terminal/C-terminal Construct of Surfactant Protein B Using 13C-Enhanced Fourier Transform Infrared (FTIR) Spectroscopy | | Descriptor: | Pulmonary surfactant-associated protein B | | Authors: | Waring, A.J, Walther, F.J, Gordon, L.M, Hernandez-Juviel, J.M, Hong, T, Sherman, M.A, Alonso, C, Alig, T, Braun, A, Bacon, D, Zasadzinski, J.A. | | Deposit date: | 2004-03-24 | | Release date: | 2004-06-15 | | Last modified: | 2024-10-30 | | Method: | INFRARED SPECTROSCOPY | | Cite: | The role of charged amphipathic helices in the structure and function of surfactant protein B.
J.Pept.Res., 66, 2005
|
|
5BQF
 
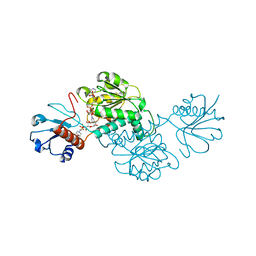 | | Probable 2-hydroxyacid dehydrogenase from Rhizobium etli CFN 42 in complex with NADP, HEPES and L(+)-tartaric acid | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Langner, K.M, Shabalin, I.G, Handing, K.B, Gasiorowska, O.A, Stead, M, Hillerich, B.S, Chowdhury, S, Hammonds, J, Zimmerman, M.D, Al Obaidi, N, Bonanno, J, Seidel, R, Almo, S.C, Minor, W, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2015-05-29 | | Release date: | 2015-06-17 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Probable 2-hydroxyacid dehydrogenase from Rhizobium etli CFN 42 in complex with NADP, HEPES and L-tartaric acid
to be published
|
|
1L6W
 
 | | Fructose-6-phosphate aldolase | | Descriptor: | Fructose-6-phosphate aldolase 1, GLYCEROL | | Authors: | Thorell, S, Schuermann, M, Sprenger, G.A, Schneider, G. | | Deposit date: | 2002-03-14 | | Release date: | 2002-06-12 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Crystal structure of decameric fructose-6-phosphate aldolase from Escherichia coli reveals inter-subunit helix swapping as a structural basis for assembly differences in the transaldolase family.
J.Mol.Biol., 319, 2002
|
|
2D09
 
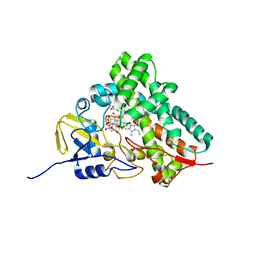 | |
2FGC
 
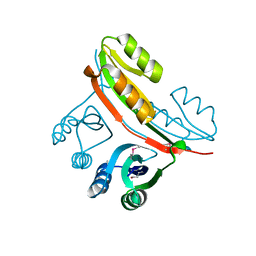 | | Crystal structure of Acetolactate synthase- small subunit from Thermotoga maritima | | Descriptor: | MAGNESIUM ION, acetolactate synthase, small subunit | | Authors: | Petkowski, J.J, Chruszcz, M, Zimmerman, M.D, Zheng, H, Cymborowski, M.T, Koclega, K.D, Kudritska, M, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2005-12-21 | | Release date: | 2006-02-07 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structures of TM0549 and NE1324--two orthologs of E. coli AHAS isozyme III small regulatory subunit.
Protein Sci., 16, 2007
|
|
3BHD
 
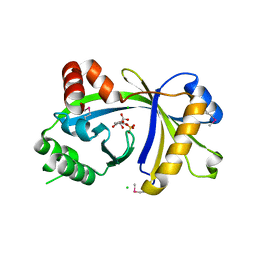 | | Crystal structure of human thiamine triphosphatase (THTPA) | | Descriptor: | CHLORIDE ION, CITRIC ACID, GLYCEROL, ... | | Authors: | Busam, R.D, Lehtio, L, Arrowsmith, C.H, Collins, R, Dahlgren, L.G, Edwards, A.M, Flodin, S, Flores, A, Graslund, S, Hammarstrom, M, Hallberg, B.M, Herman, M.D, Johansson, A, Johansson, I, Kallas, A, Karlberg, T, Kotenyova, T, Moche, M, Nilsson, M.E, Nordlund, P, Nyman, T, Persson, C, Sagemark, J, Sundstrom, M, Svensson, L, Thorsell, A.G, Tresaugues, L, Van den Berg, S, Weigelt, J, Welin, M, Berglund, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-11-28 | | Release date: | 2007-12-11 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal Structure of Human Thiamine Triphosphatase.
To be Published
|
|
3KSW
 
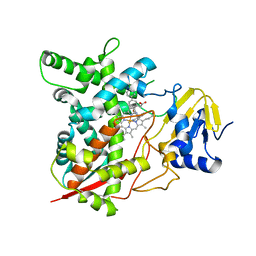 | | Crystal structure of sterol 14alpha-demethylase (CYP51) from Trypanosoma cruzi in complex with an inhibitor VNF ((4-(4-chlorophenyl)-N-[2-(1H-imidazol-1-yl)-1-phenylethyl]benzamide) | | Descriptor: | 4'-chloro-N-[(1R)-2-(1H-imidazol-1-yl)-1-phenylethyl]biphenyl-4-carboxamide, PROTOPORPHYRIN IX CONTAINING FE, Sterol 14-alpha demethylase | | Authors: | Lepesheva, G.I, Hargrove, T.Y, Anderson, S, Wawrzak, Z, Waterman, M.R. | | Deposit date: | 2009-11-23 | | Release date: | 2009-12-01 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Structural Insights into Inhibition of Sterol 14{alpha}-Demethylase in the Human Pathogen Trypanosoma cruzi.
J.Biol.Chem., 285, 2010
|
|
2QQ2
 
 | | Crystal structure of C-terminal domain of Human acyl-CoA thioesterase 7 | | Descriptor: | Cytosolic acyl coenzyme A thioester hydrolase | | Authors: | Busam, R, Lehtio, L, Arrowsmith, C.H, Berglund, H, Collins, R, Dahlgren, L.G, Herman, M.D, Edwards, A, Flodin, S, Flores, A, Graslund, S, Hammarstrom, M, Hallberg, B.M, Holmberg-Schiavone, L, Johansson, I, Kallas, A, Karlberg, T, Kotenyova, T, Moche, M, Nordlund, P, Nyman, T, Sagemark, J, Stenmark, P, Sundstrom, M, Thorsell, A.G, Tresaugues, L, van den Berg, S, Weigelt, J, Welin, M, Persson, C, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-07-26 | | Release date: | 2007-08-14 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Human acyl-CoA thioesterase 7.
To be Published
|
|
3P0L
 
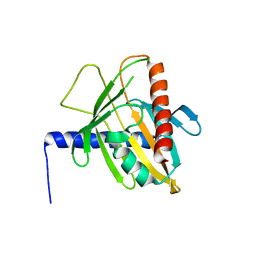 | | Human steroidogenic acute regulatory protein | | Descriptor: | Steroidogenic acute regulatory protein, mitochondrial | | Authors: | Lehtio, L, Siponen, M, Arrowsmith, C.H, Berglund, H, Busam, R.D, Collins, R, Dahlgren, L.G, Edwards, A.M, Flodin, S, Flores, A, Graslund, S, Hammarstrom, M, Hallberg, B.M, Herman, M.D, Johansson, A, Johansson, I, Kallas, A, Karlberg, T, Kotenyova, T, Moche, M, Nilsson, M.E, Nordlund, P, Nyman, T, Persson, C, Sagemark, J, Sundstrom, M, Svensson, L, Thorsell, A.G, Tresaugues, L, Van Den Berg, S, Weigelt, J, Welin, M, Schuler, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-09-29 | | Release date: | 2010-11-24 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Comparative structural analysis of lipid binding START domains.
Plos One, 6, 2011
|
|
3GQS
 
 | | Crystal structure of the FHA domain of CT664 protein from Chlamydia trachomatis | | Descriptor: | Adenylate cyclase-like protein, PHOSPHATE ION | | Authors: | Majorek, K.A, Cymborowski, M, Chruszcz, M, Evdokimova, E, Egorova, O, Di Leo, R, Zimmerman, M.D, Savchenko, A, Joachimiak, A, Edwards, A.M, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-03-24 | | Release date: | 2009-04-07 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of the FHA domain of CT664 protein from Chlamydia trachomatis
To be Published
|
|
3HFR
 
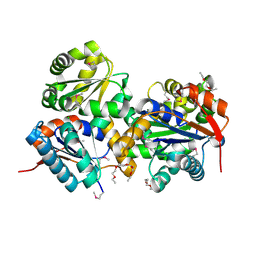 | | Crystal structure of glutamate racemase from Listeria monocytogenes | | Descriptor: | CHLORIDE ION, Glutamate racemase, O-(O-(2-AMINOPROPYL)-O'-(2-METHOXYETHYL)POLYPROPYLENE GLYCOL 500) | | Authors: | Majorek, K.A, Chruszcz, M, Zimmerman, M.D, Klimecka, M.M, Cymborowski, M, Skarina, T, Onopriyenko, O, Stam, J, Otwinowski, Z, Anderson, W.F, Savchenko, A, Minor, W, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2009-05-12 | | Release date: | 2009-06-09 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of glutamate racemase from Listeria monocytogenes
TO BE PUBLISHED
|
|
2BPO
 
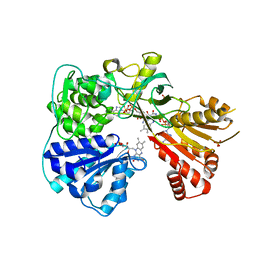 | | Crystal structure of the yeast CPR triple mutant: D74G, Y75F, K78A. | | Descriptor: | FLAVIN MONONUCLEOTIDE, FLAVIN-ADENINE DINUCLEOTIDE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Yermalitskaya, L.V, Kim, Y, Waterman, M.R, Podust, L.M. | | Deposit date: | 2005-04-21 | | Release date: | 2006-07-13 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal Structure of the Yeast Cpr Triple Mutant: D74G, Y75F, K78A.
To be Published
|
|
1S0T
 
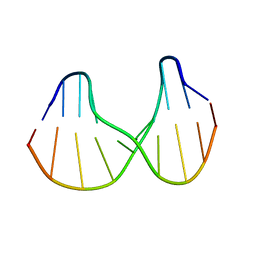 | | Solution structure of a DNA duplex containing an alpha-anomeric adenosine: insights into substrate recognition by endonuclease IV | | Descriptor: | 5'-D(*Cp*Gp*Tp*Cp*Gp*Tp*Gp*Gp*Ap*C)-3', 5'-D(*Gp*Tp*Cp*Cp*(A3A)p*Cp*Gp*Ap*Cp*G)-3' | | Authors: | Aramini, J.M, Cleaver, S.H, Pon, R.T, Cunningham, R.P, Germann, M.W. | | Deposit date: | 2004-01-04 | | Release date: | 2004-04-20 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of a DNA Duplex Containing an alpha-Anomeric Adenosine: Insights into Substrate Recognition by Endonuclease IV.
J.Mol.Biol., 338, 2004
|
|
4JYJ
 
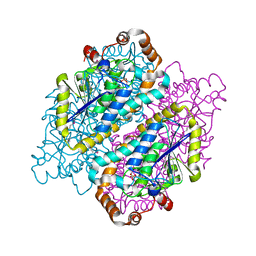 | | Crystal Structure of putative enoyl-CoA hydratase/isomerase from Novosphingobium aromaticivorans DSM 12444 | | Descriptor: | Enoyl-CoA hydratase/isomerase, UNKNOWN LIGAND | | Authors: | Cooper, D.R, Porebski, P.J, Domagalski, M.J, Ahmed, M, Stead, M, Hillerich, B, Seidel, R, Zimmerman, M, Bonanno, J.B, Almo, S.C, Minor, W, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2013-03-29 | | Release date: | 2013-05-29 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structure of putative enoyl-CoA hydratase/isomerase from Novosphingobium aromaticivorans DSM 12444
To be Published
|
|
2BF4
 
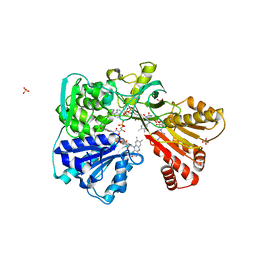 | | A second FMN-binding site in yeast NADPH-cytochrome P450 reductase suggests a novel mechanism of electron transfer by diflavin reductases. | | Descriptor: | FLAVIN MONONUCLEOTIDE, FLAVIN-ADENINE DINUCLEOTIDE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Podust, L.M, Lepesheva, G.I, Kim, Y, Yermalitskaya, L.V, Yermalitsky, V.N, Lamb, D.C, Kelly, S.L, Waterman, M.R. | | Deposit date: | 2004-12-03 | | Release date: | 2006-01-17 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | A Second Fmn-Binding Site in Yeast Nadph-Cytochrome P450 Reductase Suggests a Mechanism of Electron Transfer by Diflavin Reductases.
Structure, 14, 2006
|
|
3POP
 
 | | The crystal structure of GilR, an oxidoreductase that catalyzes the terminal step of gilvocarcin biosynthesis | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, GilR oxidase | | Authors: | Noinaj, N, Bosserman, M.A, Schickli, M.A, Kharel, M.K, Rohr, J, Buchanan, S.K. | | Deposit date: | 2010-11-23 | | Release date: | 2011-05-11 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.651 Å) | | Cite: | The Crystal Structure and Mechanism of an Unusual Oxidoreductase, GilR, Involved in Gilvocarcin V Biosynthesis.
J.Biol.Chem., 286, 2011
|
|
3III
 
 | | 1.95 Angstrom Crystal Structure of CocE/NonD family hydrolase (SACOL2612) from Staphylococcus aureus | | Descriptor: | CHLORIDE ION, CocE/NonD family hydrolase, NICKEL (II) ION, ... | | Authors: | Osinski, T, Chruszcz, M, Domagalski, M.J, Cymborowski, M, Shumilin, I.A, Skarina, T, Onopriyenko, O, Zimmerman, M.D, Savchenko, A, Edwards, A, Anderson, W.F, Minor, W, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2009-08-01 | | Release date: | 2009-08-18 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | 1.95 Angstrom Crystal Structure of CocE/NonD family hydrolase (SACOL2612) from Staphylococcus aureus
To be Published
|
|
