1D5T
 
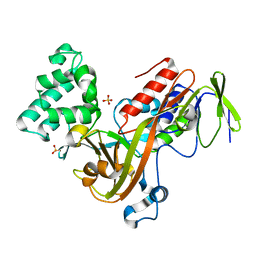 | | GUANINE NUCLEOTIDE DISSOCIATION INHIBITOR, ALPHA-ISOFORM | | Descriptor: | GUANINE NUCLEOTIDE DISSOCIATION INHIBITOR, SULFATE ION | | Authors: | Peng, L, Zeng, K, Heine, A, Moyer, B, Greasley, S.E, Kuhn, P, Balch, W.E, Wilson, I.A. | | Deposit date: | 1999-10-11 | | Release date: | 2000-10-25 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.04 Å) | | Cite: | A new functional domain of guanine nucleotide dissociation inhibitor (alpha-GDI) involved in Rab recycling.
Traffic, 1, 2000
|
|
5F5I
 
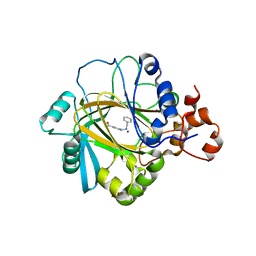 | | Crystal Structure of human JMJD2A complexed with KDOOA011340 | | Descriptor: | 2-[[(phenylmethyl)amino]methyl]pyridine-4-carboxylic acid, Lysine-specific demethylase 4A, NICKEL (II) ION, ... | | Authors: | Krojer, T, Vollmar, M, Crawley, L, Szykowska, A, Gileadi, C, Johansson, C, England, K, Yang, H, Burgess-Brown, N, Brennan, P, Bountra, C, Arrowsmith, C.H, Edwards, A, Oppermann, U, von Delft, F, Structural Genomics Consortium (SGC) | | Deposit date: | 2015-12-04 | | Release date: | 2015-12-30 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.63 Å) | | Cite: | 8-Substituted Pyrido[3,4-d]pyrimidin-4(3H)-one Derivatives As Potent, Cell Permeable, KDM4 (JMJD2) and KDM5 (JARID1) Histone Lysine Demethylase Inhibitors.
J.Med.Chem., 59, 2016
|
|
2MG9
 
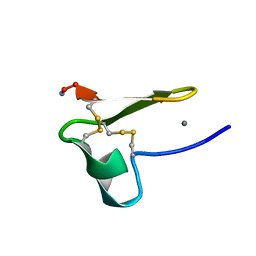 | | Truncated EGF-A | | Descriptor: | CALCIUM ION, Low-density lipoprotein receptor | | Authors: | Schroeder, C.I, Rosengren, K. | | Deposit date: | 2013-10-30 | | Release date: | 2014-04-02 | | Last modified: | 2024-11-06 | | Method: | SOLUTION NMR | | Cite: | Design and Synthesis of Truncated EGF-A Peptides that Restore LDL-R Recycling in the Presence of PCSK9 In Vitro.
Chem.Biol., 21, 2014
|
|
4GUX
 
 | | Crystal structure of trypsin:MCoTi-II complex | | Descriptor: | ACETATE ION, CALCIUM ION, Cationic trypsin, ... | | Authors: | King, G.J, Daly, N.L, Thorstholm, L, Greenwood, K.P, Rosengren, K.J, Heras, B, Craik, D.J, Martin, J.L. | | Deposit date: | 2012-08-30 | | Release date: | 2013-09-04 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.803 Å) | | Cite: | Structural insights into the role of the cyclic backbone in a squash trypsin inhibitor
J.Biol.Chem., 288, 2013
|
|
5F55
 
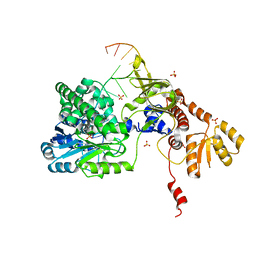 | | Structure of RecJ complexed with DNA | | Descriptor: | DNA (5'-D(*GP*AP*TP*GP*TP*AP*CP*GP*CP*TP*AP*GP*GP*C)-3'), MANGANESE (II) ION, SULFATE ION, ... | | Authors: | Hua, Y, Zhao, Y, Cheng, K. | | Deposit date: | 2015-12-04 | | Release date: | 2016-06-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural basis for DNA 5 -end resection by RecJ
Elife, 5, 2016
|
|
5F56
 
 | | Structure of RecJ complexed with DNA and SSB-ct | | Descriptor: | ALA-ASP-LEU-PRO-PHE, DNA (5'-D(*CP*TP*GP*AP*TP*GP*GP*CP*A)-3'), MANGANESE (II) ION, ... | | Authors: | Zhao, Y, Hua, Y, Cheng, K. | | Deposit date: | 2015-12-04 | | Release date: | 2016-06-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis for DNA 5 -end resection by RecJ
Elife, 5, 2016
|
|
3B2R
 
 | | Crystal Structure of PDE5A1 catalytic domain in complex with Vardenafil | | Descriptor: | 2-{2-ETHOXY-5-[(4-ETHYLPIPERAZIN-1-YL)SULFONYL]PHENYL}-5-METHYL-7-PROPYLIMIDAZO[5,1-F][1,2,4]TRIAZIN-4(1H)-ONE, cGMP-specific 3',5'-cyclic phosphodiesterase | | Authors: | Huanchen, W, Mengchun, Y, Howard, R, Sharron, H.F, Hengming, K. | | Deposit date: | 2007-10-19 | | Release date: | 2008-05-20 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Conformational variations of both phosphodiesterase-5 and inhibitors provide the structural basis for the physiological effects of vardenafil and sildenafil.
Mol.Pharmacol., 73, 2008
|
|
2LWV
 
 | |
2LWT
 
 | |
4GVJ
 
 | | Tyk2 (JH1) in complex with adenosine di-phosphate | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Non-receptor tyrosine-protein kinase TYK2 | | Authors: | Liang, J, Abbema, A.V, Bao, L, Barrett, K, Beresini, M, Berezhkovskiy, L, Blair, W, Chang, C, Driscoll, J, Eigenbrot, C, Ghilardi, N, Gibbons, P, Halladay, J, Johnson, A, Kohli, P.B, Lai, Y, Liimatta, M, Mantik, P, Menghrajani, K, Murray, J, Sambrone, A, Shao, Y, Shia, S, Shin, Y, Smith, J, Sohn, S, Stanley, M, Tsui, V, Ultsch, M, Wu, L, Zhang, B, Magnuson, S. | | Deposit date: | 2012-08-30 | | Release date: | 2013-08-14 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Lead identification of novel and selective TYK2 inhibitors.
Eur.J.Med.Chem., 67, 2013
|
|
2LWS
 
 | |
2LWU
 
 | |
2LWQ
 
 | |
5F54
 
 | | Structure of RecJ complexed with dTMP | | Descriptor: | MANGANESE (II) ION, Single-stranded-DNA-specific exonuclease, THYMIDINE-5'-PHOSPHATE | | Authors: | Hua, Y, Zhao, Y, Cheng, K. | | Deposit date: | 2015-12-04 | | Release date: | 2016-06-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis for DNA 5 -end resection by RecJ
Elife, 5, 2016
|
|
3DEP
 
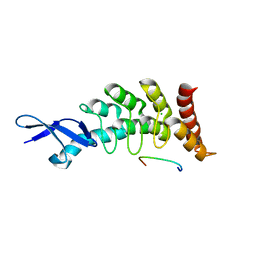 | | Structural basis for specific substrate recognition by the chloroplast signal recognition particle protein cpSRP43 | | Descriptor: | CHLORIDE ION, Signal recognition particle 43 kDa protein, YPGGSFDPLGLA | | Authors: | Holdermann, I, Stengel, K.F, Wild, K, Sinning, I. | | Deposit date: | 2008-06-10 | | Release date: | 2008-08-12 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis for specific substrate recognition by the chloroplast signal recognition particle protein cpSRP43.
Science, 321, 2008
|
|
2JIU
 
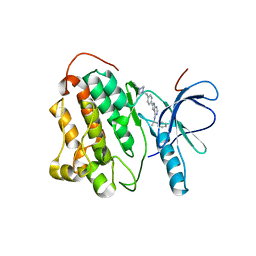 | | Crystal structure of EGFR kinase domain T790M mutation in complex with AEE788 | | Descriptor: | 6-{4-[(4-ETHYLPIPERAZIN-1-YL)METHYL]PHENYL}-N-[(1R)-1-PHENYLETHYL]-7H-PYRROLO[2,3-D]PYRIMIDIN-4-AMINE, EPIDERMAL GROWTH FACTOR RECEPTOR | | Authors: | Yun, C.-H, Mengwasser, K.E, Toms, A.V, Woo, M.S, Greulich, H, Wong, K.-K, Meyerson, M, Eck, M.J. | | Deposit date: | 2007-07-01 | | Release date: | 2008-01-22 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | The T790M Mutation in Egfr Kinase Causes Drug Resistance by Increasing the Affinity for ATP.
Proc.Natl.Acad.Sci.USA, 105, 2008
|
|
2JIT
 
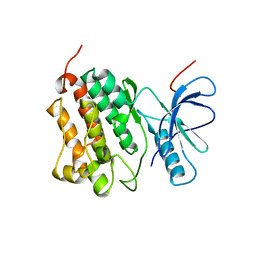 | | Crystal structure of EGFR kinase domain T790M mutation | | Descriptor: | EPIDERMAL GROWTH FACTOR RECEPTOR | | Authors: | Yun, C.-H, Mengwasser, K.E, Toms, A.V, Woo, M.S, Greulich, H, Wong, K.-K, Meyerson, M, Eck, M.J. | | Deposit date: | 2007-07-01 | | Release date: | 2008-01-22 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | The T790M Mutation in Egfr Kinase Causes Drug Resistance by Increasing the Affinity for ATP.
Proc.Natl.Acad.Sci.USA, 105, 2008
|
|
2JIV
 
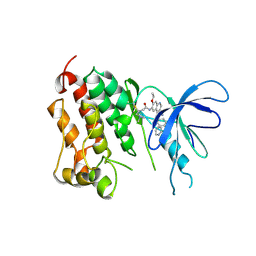 | | Crystal structure of EGFR kinase domain T790M mutation in compex with HKI-272 | | Descriptor: | CHLORIDE ION, EPIDERMAL GROWTH FACTOR RECEPTOR, N-(4-{[3-chloro-4-(pyridin-2-ylmethoxy)phenyl]amino}-3-cyano-7-ethoxyquinolin-6-yl)-4-(dimethylamino)butanamide | | Authors: | Yun, C.-H, Mengwasser, K.E, Toms, A.V, Li, Y, Woo, M.S, Greulich, H, Wong, K.-K, Meyerson, M, Eck, M.J. | | Deposit date: | 2007-07-02 | | Release date: | 2008-01-22 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | The T790M Mutation in Egfr Kinase Causes Drug Resistance by Increasing the Affinity for ATP.
Proc.Natl.Acad.Sci.USA, 105, 2008
|
|
1ZXF
 
 | | Solution structure of a self-sacrificing resistance protein, CalC from Micromonospora echinospora | | Descriptor: | CalC | | Authors: | Singh, S, Hager, M.H, Zhang, C, Griffith, B.R, Lee, M.S, Hallenga, K, Markley, J.L, Thorson, J.S, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2005-06-08 | | Release date: | 2005-12-13 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structural insight into the self-sacrifice mechanism of enediyne resistance.
Acs Chem.Biol., 1, 2006
|
|
1ZA8
 
 | | NMR solution structure of a leaf-specific-expressed cyclotide vhl-1 | | Descriptor: | vhl-1 | | Authors: | Chen, B, Colgrave, M.L, Daly, N.L, Rosengren, K.J, Gustafson, K.R, Craik, D.J. | | Deposit date: | 2005-04-05 | | Release date: | 2005-04-12 | | Last modified: | 2024-10-23 | | Method: | SOLUTION NMR | | Cite: | Isolation and characterization of novel cyclotides from Viola hederaceae: solution structure and anti-HIV activity of vhl-1, a leaf-specific expressed cyclotide.
J.Biol.Chem., 280, 2005
|
|
2ATG
 
 | | NMR structure of Retrocyclin-2 in SDS | | Descriptor: | Retrocyclin-2 | | Authors: | Daly, N.L, Chen, Y.K, Rosengren, K.J, Marx, U.C, Phillips, M.L, Waring, A.J, Wang, W, Lehrer, R.I, Craik, D.J. | | Deposit date: | 2005-08-24 | | Release date: | 2005-09-06 | | Last modified: | 2024-11-13 | | Method: | SOLUTION NMR | | Cite: | Retrocyclin-2: structural analysis of a potent anti-HIV theta-defensin
Biochemistry, 46, 2007
|
|
2M2S
 
 | | Solution structure of the antimicrobial peptide [Aba5,7,12,14]BTD-2 | | Descriptor: | [Aba5,7,12,14]BTD-2 | | Authors: | Conibear, A.C, Rosengren, K, Daly, N.L, Troiera Henriques, S, Craik, D.J. | | Deposit date: | 2013-01-02 | | Release date: | 2013-02-27 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | The cyclic cystine ladder in theta-defensins is important for structure and stability, but not antibacterial activity.
J.Biol.Chem., 288, 2013
|
|
2M2Y
 
 | | Solution structure of the antimicrobial peptide Btd-2[3,4] | | Descriptor: | BTD-2[3,4] | | Authors: | Conibear, A.C, Rosengren, K, Daly, N.L, Troiera Henriques, S, Craik, D.J. | | Deposit date: | 2013-01-03 | | Release date: | 2013-02-27 | | Last modified: | 2024-10-09 | | Method: | SOLUTION NMR | | Cite: | The cyclic cystine ladder in theta-defensins is important for structure and stability, but not antibacterial activity.
J.Biol.Chem., 288, 2013
|
|
2M77
 
 | | [Asp2]RTD-1 | | Descriptor: | [Asp2]RTD-1 | | Authors: | Conibear, A.C, Bochen, A, Rosengren, K, Kessler, H, Craik, D.J. | | Deposit date: | 2013-04-18 | | Release date: | 2014-02-05 | | Last modified: | 2024-11-20 | | Method: | SOLUTION NMR | | Cite: | The Cyclic Cystine Ladder of Theta-Defensins as a Stable, Bifunctional Scaffold: A Proof-of-Concept Study Using the Integrin-Binding RGD Motif
Chembiochem, 15, 2014
|
|
2LYF
 
 | |
