2J1D
 
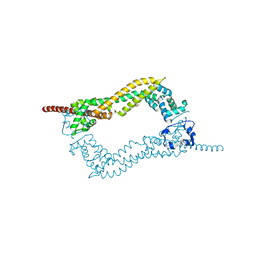 | | Crystallization of hDaam1 C-terminal Fragment | | Descriptor: | DISHEVELED-ASSOCIATED ACTIVATOR OF MORPHOGENESIS 1, GLYCEROL, PHOSPHATE ION | | Authors: | Lu, J, Meng, W, Poy, F, Eck, M.J. | | Deposit date: | 2006-08-10 | | Release date: | 2007-05-15 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structure of the Fh2 Domain of Daam1: Implications for Formin Regulation of Actin Assembly.
J.Mol.Biol., 369, 2007
|
|
2SRC
 
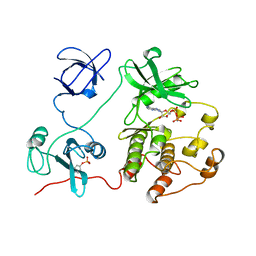 | | CRYSTAL STRUCTURE OF HUMAN TYROSINE-PROTEIN KINASE C-SRC, IN COMPLEX WITH AMP-PNP | | Descriptor: | PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, TYROSINE-PROTEIN KINASE SRC | | Authors: | Xu, W, Doshi, A, Lei, M, Eck, M.J, Harrison, S.C. | | Deposit date: | 1998-12-29 | | Release date: | 1999-07-22 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structures of c-Src reveal features of its autoinhibitory mechanism.
Mol.Cell, 3, 1999
|
|
2IUH
 
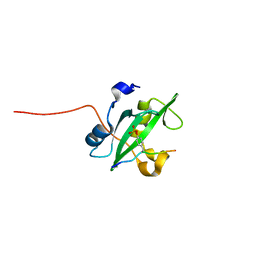 | | Crystal structure of the PI3-kinase p85 N-terminal SH2 domain in complex with c-Kit phosphotyrosyl peptide | | Descriptor: | C-KIT PHOSPHOTYROSYL PEPTIDE, PHOSPHATIDYLINOSITOL 3-KINASE REGULATORY ALPHA SUBUNIT | | Authors: | Nolte, R.T, Eck, M.J, Schlessinger, J, Shoelson, S.E, Harrison, S.C. | | Deposit date: | 2006-06-03 | | Release date: | 2006-06-06 | | Last modified: | 2021-04-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of the Pi 3-Kinase P85 Amino-Terminal Sh2 Domain and its Phosphopeptide Complexes
Nat.Struct.Biol., 3, 1996
|
|
2JKK
 
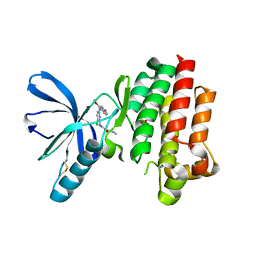 | |
2SHP
 
 | | TYROSINE PHOSPHATASE SHP-2 | | Descriptor: | DODECANE-TRIMETHYLAMINE, SHP-2 | | Authors: | Hof, P, Pluskey, S, Dhe-Paganon, S, Eck, M.J, Shoelson, S.E. | | Deposit date: | 1997-12-01 | | Release date: | 1999-02-16 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of the tyrosine phosphatase SHP-2.
Cell(Cambridge,Mass.), 92, 1998
|
|
1KSW
 
 | | Structure of Human c-Src Tyrosine Kinase (Thr338Gly Mutant) in Complex with N6-benzyl ADP | | Descriptor: | N6-BENZYL ADENOSINE-5'-DIPHOSPHATE, PROTO-ONCOGENE TYROSINE-PROTEIN KINASE SRC | | Authors: | Witucki, L.A, Huang, X, Shah, K, Liu, Y, Kyin, S, Eck, M.J, Shokat, K.M. | | Deposit date: | 2002-01-14 | | Release date: | 2002-02-27 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Mutant tyrosine kinases with unnatural nucleotide specificity retain the structure and phospho-acceptor specificity of the wild-type enzyme.
Chem.Biol., 9, 2002
|
|
1L3E
 
 | | NMR Structures of the HIF-1alpha CTAD/p300 CH1 Complex | | Descriptor: | ZINC ION, hypoxia inducible factor-1 alpha subunit, p300 protein | | Authors: | Freedman, S.J, Sun, Z.J, Poy, F, Kung, A.L, Livingston, D.M, Wagner, G, Eck, M.J. | | Deposit date: | 2002-02-26 | | Release date: | 2002-04-24 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Structural basis for recruitment of CBP/p300 by hypoxia-inducible factor-1 alpha.
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
1JPW
 
 | | Crystal Structure of a Human Tcf-4 / beta-Catenin Complex | | Descriptor: | BETA-CATENIN, transcription factor 7-like 2 | | Authors: | Poy, F, Lepourcelet, M, Shivdasani, R.A, Eck, M.J. | | Deposit date: | 2001-08-03 | | Release date: | 2001-12-05 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of a human Tcf4-beta-catenin complex.
Nat.Struct.Biol., 8, 2001
|
|
1M27
 
 | | Crystal structure of SAP/FynSH3/SLAM ternary complex | | Descriptor: | CITRATE ANION, Proto-oncogene tyrosine-protein kinase FYN, SH2 domain protein 1A, ... | | Authors: | Chan, B, Griesbach, J, Song, H.K, Poy, F, Terhorst, C, Eck, M.J. | | Deposit date: | 2002-06-21 | | Release date: | 2003-05-06 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | SAP couples Fyn to SLAM immune receptors.
NAT.CELL BIOL., 5, 2003
|
|
1IJQ
 
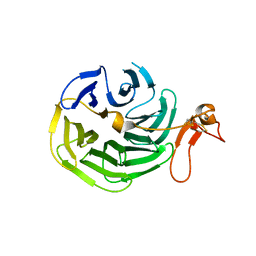 | | Crystal Structure of the LDL Receptor YWTD-EGF Domain Pair | | Descriptor: | LOW-DENSITY LIPOPROTEIN RECEPTOR | | Authors: | Jeon, H, Meng, W, Takagi, J, Eck, M.J, Springer, T.A, Blacklow, S.C. | | Deposit date: | 2001-04-27 | | Release date: | 2001-05-23 | | Last modified: | 2018-04-04 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Implications for familial hypercholesterolemia from the structure of the LDL receptor YWTD-EGF domain pair.
Nat.Struct.Biol., 8, 2001
|
|
8BGU
 
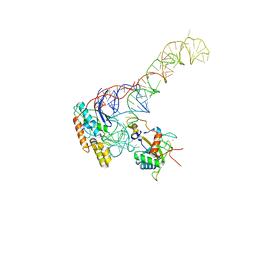 | | human MDM2-5S RNP | | Descriptor: | 5S rRNA, 60S ribosomal protein L11, 60S ribosomal protein L5, ... | | Authors: | Castillo, N, Thoms, M, Flemming, D, Hammaren, H.M, Buschauer, R, Ameismeier, M, Bassler, J, Beck, M, Beckmann, R, Hurt, E. | | Deposit date: | 2022-10-28 | | Release date: | 2023-06-14 | | Last modified: | 2023-08-30 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Structure of nascent 5S RNPs at the crossroad between ribosome assembly and MDM2-p53 pathways.
Nat.Struct.Mol.Biol., 30, 2023
|
|
1W8K
 
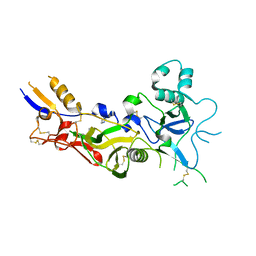 | | Crystal structure of apical membrane antigen 1 from Plasmodium vivax | | Descriptor: | APICAL MEMBRANE ANTIGEN 1, IMIDAZOLE | | Authors: | Pizarro, J.C, Vulliez-Le Normand, B, Chesne-Seck, M.-L, Kocken, C.H.M, Thomas, A.W, Bentley, G.A. | | Deposit date: | 2004-09-23 | | Release date: | 2005-03-01 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of the Malaria Vaccine Candidate Apical Membrane Antigen 1
Science, 308, 2005
|
|
4FBL
 
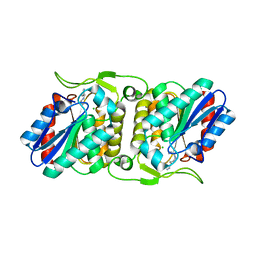 | | LipS and LipT, two metagenome-derived lipolytic enzymes increase the diversity of known lipase and esterase families | | Descriptor: | CHLORIDE ION, LipS lipolytic enzyme, SPERMIDINE | | Authors: | Chow, J, Krauss, U, Dall Antonia, Y, Fersini, F, Schmeisser, C, Schmidt, M, Menyes, I, Bornscheuer, U, Lauinger, B, Bongen, P, Pietruszka, J, Eckstein, M, Thum, O, Liese, A, Mueller-Dieckmann, J, Jaeger, K.-E, Kovacic, F, Streit, W.R, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2012-05-23 | | Release date: | 2012-10-10 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | The Metagenome-Derived Enzymes LipS and LipT Increase the Diversity of Known Lipases.
Plos One, 7, 2012
|
|
8S03
 
 | | NMR solution structure of the CysD2 domain of MUC2 | | Descriptor: | CALCIUM ION, Mucin-2 | | Authors: | Recktenwald, C, Karlsson, B.G, Garcia-Bonnete, M.-J, Katona, G, Jensen, M, Lymer, R, Baeckstroem, M, Johansson, M.E.V, Hansson, G.C, Trillo-Muyo, S. | | Deposit date: | 2024-02-13 | | Release date: | 2024-04-17 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The structure of the second CysD domain of MUC2 and role in mucin organization by transglutaminase-based cross-linking.
Cell Rep, 43, 2024
|
|
4FBM
 
 | | LipS and LipT, two metagenome-derived lipolytic enzymes increase the diversity of known lipase and esterase families | | Descriptor: | BROMIDE ION, LipS lipolytic enzyme | | Authors: | Chow, J, Krauss, U, Dall Antonia, Y, Fersini, F, Schmeisser, C, Schmidt, M, Menyes, I, Bornscheuer, U, Lauinger, B, Bongen, P, Pietruszka, J, Eckstein, M, Thum, O, Liese, A, Mueller-Dieckmann, J, Jaeger, K.-E, Kovavic, F, Streit, W.R, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2012-05-23 | | Release date: | 2012-10-10 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The Metagenome-Derived Enzymes LipS and LipT Increase the Diversity of Known Lipases.
Plos One, 7, 2012
|
|
4V6I
 
 | | Localization of the small subunit ribosomal proteins into a 6.1 A cryo-EM map of Saccharomyces cerevisiae translating 80S ribosome | | Descriptor: | 18S rRNA, 25S rRNA, 40S ribosomal protein RACK1 (RACK1), ... | | Authors: | Armache, J.-P, Jarasch, A, Anger, A.M, Villa, E, Becker, T, Bhushan, S, Jossinet, F, Habeck, M, Dindar, G, Franckenberg, S, Marquez, V, Mielke, T, Thomm, M, Berninghausen, O, Beatrix, B, Soeding, J, Westhof, E, Wilson, D.N, Beckmann, R. | | Deposit date: | 2010-10-12 | | Release date: | 2014-07-09 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (8.8 Å) | | Cite: | Cryo-EM structure and rRNA model of a translating eukaryotic 80S ribosome at 5.5-A resolution.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
4V7E
 
 | | Model of the small subunit RNA based on a 5.5 A cryo-EM map of Triticum aestivum translating 80S ribosome | | Descriptor: | 18S ribosomal RNA, 40S ribosomal protein S10, 40S ribosomal protein S10E, ... | | Authors: | Barrio-Garcia, C, Armache, J.-P, Jarasch, A, Anger, A.M, Villa, E, Becker, T, Bhushan, S, Jossinet, F, Habeck, M, Dindar, G, Franckenberg, S, Marquez, V, Mielke, T, Thomm, M, Berninghausen, O, Beatrix, B, Soeding, J, Westhof, E, Wilson, D.N, Beckmann, R. | | Deposit date: | 2013-11-22 | | Release date: | 2014-07-09 | | Last modified: | 2023-02-01 | | Method: | ELECTRON MICROSCOPY (5.5 Å) | | Cite: | Structures of the Sec61 complex engaged in nascent peptide translocation or membrane insertion.
Nature, 506, 2014
|
|
2XIU
 
 | | High resolution structure of MTSL-tagged CylR2. | | Descriptor: | CYLR2, GLYCEROL, S-[(1-oxyl-2,2,5,5-tetramethyl-2,5-dihydro-1H-pyrrol-3-yl)methyl] methanesulfonothioate | | Authors: | Gruene, T, Cho, M.-K, Karyagina, I, Kim, H.-Y, Grosse, C, Giller, K, Zweckstetter, M, Becker, S. | | Deposit date: | 2010-07-01 | | Release date: | 2011-02-09 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Integrated Analysis of the Conformation of a Protein-Linked Spin Label by Crystallography, Epr and NMR Spectroscopy.
J.Biomol.NMR, 49, 2011
|
|
2XJ3
 
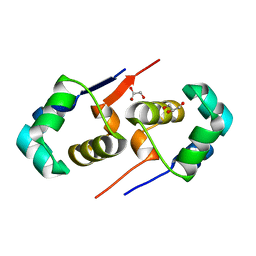 | | High resolution structure of the T55C mutant of CylR2. | | Descriptor: | CYLR2 SYNONYM CYTOLYSIN REPRESSOR 2, GLYCEROL | | Authors: | Gruene, T, Cho, M.K, Karyagina, I, Kim, H.Y, Grosse, C, Giller, K, Zweckstetter, M, Becker, S. | | Deposit date: | 2010-07-02 | | Release date: | 2011-02-09 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.23 Å) | | Cite: | Integrated Analysis of the Conformation of a Protein-Linked Spin Label by Crystallography, Epr and NMR Spectroscopy.
J.Biomol.NMR, 49, 2011
|
|
4V6X
 
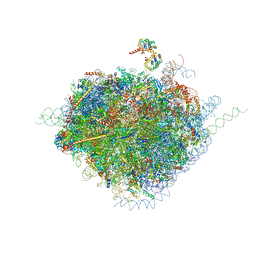 | | Structure of the human 80S ribosome | | Descriptor: | 18S ribosomal RNA, 28S ribosomal RNA, 40S ribosomal protein S10, ... | | Authors: | Anger, A.M, Armache, J.-P, Berninghausen, O, Habeck, M, Subklewe, M, Wilson, D.N, Beckmann, R. | | Deposit date: | 2013-02-27 | | Release date: | 2014-07-09 | | Last modified: | 2023-02-01 | | Method: | ELECTRON MICROSCOPY (5 Å) | | Cite: | Structures of the human and Drosophila 80S ribosome.
Nature, 497, 2013
|
|
6R2H
 
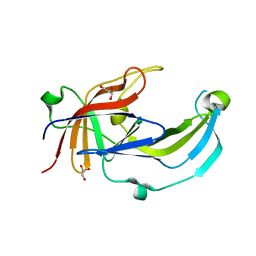 | | Crystal structure of Apo PinO from Porphyromonas gingivitis | | Descriptor: | GLYCEROL, HmuY protein | | Authors: | Antonyuk, S.V, Bielecki, M, Strange, R.W, Capper, M, Olczak, T, Olczak, M. | | Deposit date: | 2019-03-17 | | Release date: | 2020-01-15 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | Prevotella intermedia produces two proteins homologous to Porphyromonas gingivalis HmuY but with different heme coordination mode.
Biochem.J., 477, 2020
|
|
8S32
 
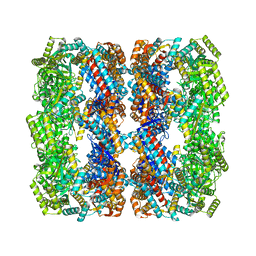 | | GroEL with bound GroTAC peptide | | Descriptor: | Chaperonin GroEL, GroTAC | | Authors: | Wroblewski, K, Izert-Nowakowska, M.A, Goral, T.K, Klimecka, M.M, Kmiecik, S, Gorna, M.W. | | Deposit date: | 2024-02-19 | | Release date: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (2.45 Å) | | Cite: | Depletion of essential GroEL protein in Escherichia coli using Clp-Interacting Peptidic Protein Erasers (CLIPPERs)
To Be Published
|
|
4V6W
 
 | | Structure of the D. melanogaster 80S ribosome | | Descriptor: | 18S ribosomal RNA, 28S ribosomal RNA, 2S ribosomal RNA, ... | | Authors: | Anger, A.M, Armache, J.-P, Berninghausen, O, Habeck, M, Subklewe, M, Wilson, D.N, Beckmann, R. | | Deposit date: | 2013-02-27 | | Release date: | 2014-07-09 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (6 Å) | | Cite: | Structures of the human and Drosophila 80S ribosome.
Nature, 497, 2013
|
|
7R5K
 
 | | Human nuclear pore complex (constricted) | | Descriptor: | Aladin, E3 SUMO-protein ligase RanBP2, Nuclear pore complex protein Nup107, ... | | Authors: | Mosalaganti, S, Obarska-Kosinska, A, Siggel, M, Taniguchi, R, Turonova, B, Zimmerli, C.E, Buczak, K, Schmidt, F.H, Margiotta, E, Mackmull, M.T, Hagen, W.J.H, Hummer, G, Kosinski, J, Beck, M. | | Deposit date: | 2022-02-10 | | Release date: | 2022-06-22 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (12 Å) | | Cite: | AI-based structure prediction empowers integrative structural analysis of human nuclear pores.
Science, 376, 2022
|
|
7R5J
 
 | | Human nuclear pore complex (dilated) | | Descriptor: | Aladin, E3 SUMO-protein ligase RanBP2, Nuclear pore complex protein Nup107, ... | | Authors: | Mosalaganti, S, Obarska-Kosinska, A, Siggel, M, Taniguchi, R, Turonova, B, Zimmerli, C.E, Buczak, K, Schmidt, F.H, Margiotta, E, Mackmull, M.T, Hagen, W.J.H, Hummer, G, Kosinski, J, Beck, M. | | Deposit date: | 2022-02-10 | | Release date: | 2022-09-21 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (50 Å) | | Cite: | AI-based structure prediction empowers integrative structural analysis of human nuclear pores
Science, 376, 2022
|
|
