6V2U
 
 | |
1UX5
 
 | | Crystal Structures of a Formin Homology-2 domain reveal a flexibly tethered dimer architecture | | Descriptor: | BNI1 PROTEIN | | Authors: | Xu, Y, Moseley, J.B, Sagot, I, Poy, F, Pellman, D, Goode, B.L, Eck, M.J. | | Deposit date: | 2004-02-19 | | Release date: | 2004-03-11 | | Last modified: | 2019-05-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structures of a Formin Homology-2 Domain Reveal a Tethered Dimer Architecture
Cell(Cambridge,Mass.), 116, 2004
|
|
7JXP
 
 | | EGFR kinase (T790M/V948R) in complex with osimertinib and JBJ-04-125-02 | | Descriptor: | (2R)-2-(5-fluoro-2-hydroxyphenyl)-2-{1-oxo-6-[4-(piperazin-1-yl)phenyl]-1,3-dihydro-2H-isoindol-2-yl}-N-(1,3-thiazol-2-yl)acetamide, Epidermal growth factor receptor, MAGNESIUM ION, ... | | Authors: | Beyett, T.S, Eck, M.J. | | Deposit date: | 2020-08-27 | | Release date: | 2021-09-08 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Molecular basis for cooperative binding and synergy of ATP-site and allosteric EGFR inhibitors
Nat Commun, 13, 2022
|
|
7JXM
 
 | | EGFR kinase (T790M/V948R) in complex with osimertinib and EAI045 | | Descriptor: | (2R)-2-(5-fluoro-2-hydroxyphenyl)-2-(1-oxo-1,3-dihydro-2H-isoindol-2-yl)-N-(1,3-thiazol-2-yl)acetamide, Epidermal growth factor receptor, MAGNESIUM ION, ... | | Authors: | Beyett, T.S, Eck, M.J. | | Deposit date: | 2020-08-27 | | Release date: | 2021-09-08 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.192 Å) | | Cite: | Molecular basis for cooperative binding and synergy of ATP-site and allosteric EGFR inhibitors
Nat Commun, 13, 2022
|
|
7JXK
 
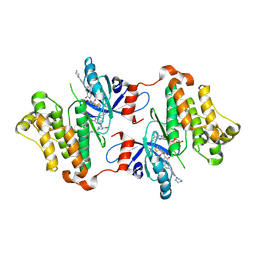 | | EGFR kinase (T790M/V948R) in complex with PF-06747775 and JBJ-04-125-02 | | Descriptor: | (2R)-2-(5-fluoro-2-hydroxyphenyl)-2-{1-oxo-6-[4-(piperazin-1-yl)phenyl]-1,3-dihydro-2H-isoindol-2-yl}-N-(1,3-thiazol-2-yl)acetamide, Epidermal growth factor receptor, MAGNESIUM ION, ... | | Authors: | Beyett, T.S, Eck, M.J. | | Deposit date: | 2020-08-27 | | Release date: | 2021-09-08 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Molecular basis for cooperative binding and synergy of ATP-site and allosteric EGFR inhibitors
Nat Commun, 13, 2022
|
|
7K1H
 
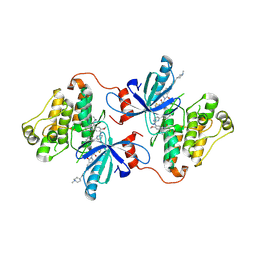 | | EGFR L858R/V948R in complex with osimertinib and allosteric inhibitor JBJ-09-063 | | Descriptor: | (2R)-2-(5-fluoro-2-hydroxyphenyl)-2-{6-[4-(1-methylpiperidin-4-yl)phenyl]-1-oxo-1,3-dihydro-2H-isoindol-2-yl}-N-(1,3-thiazol-2-yl)acetamide, Epidermal growth factor receptor, N-(2-{[2-(dimethylamino)ethyl](methyl)amino}-4-methoxy-5-{[4-(1-methyl-1H-indol-3-yl)pyrimidin-2-yl]amino}phenyl)prop-2-enamide | | Authors: | Beyett, T.S, Eck, M.J. | | Deposit date: | 2020-09-07 | | Release date: | 2021-09-15 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.602 Å) | | Cite: | Molecular basis for cooperative binding and synergy of ATP-site and allosteric EGFR inhibitors
Nat Commun, 13, 2022
|
|
7JXW
 
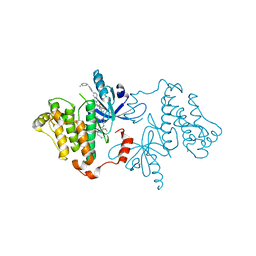 | | EGFR kinase (T790M/V948R) in complex with osimertinib and JBJ-09-063 | | Descriptor: | (2R)-2-(5-fluoro-2-hydroxyphenyl)-2-{6-[4-(1-methylpiperidin-4-yl)phenyl]-1-oxo-1,3-dihydro-2H-isoindol-2-yl}-N-(1,3-thiazol-2-yl)acetamide, Epidermal growth factor receptor, N-(2-{[2-(dimethylamino)ethyl](methyl)amino}-4-methoxy-5-{[4-(1-methyl-1H-indol-3-yl)pyrimidin-2-yl]amino}phenyl)prop-2-enamide | | Authors: | Beyett, T.S, Eck, M.J. | | Deposit date: | 2020-08-28 | | Release date: | 2021-09-08 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Molecular basis for cooperative binding and synergy of ATP-site and allosteric EGFR inhibitors
Nat Commun, 13, 2022
|
|
1U5F
 
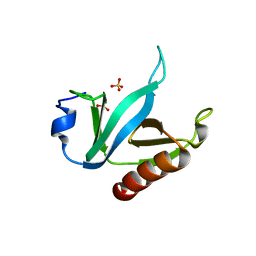 | |
1U5G
 
 | |
1B47
 
 | | STRUCTURE OF THE N-TERMINAL DOMAIN OF CBL IN COMPLEX WITH ITS BINDING SITE IN ZAP-70 | | Descriptor: | CALCIUM ION, CBL | | Authors: | Meng, W, Sawasdikosol, S, Burakoff, S.J, Eck, M.J. | | Deposit date: | 1999-01-06 | | Release date: | 1999-04-27 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of the amino-terminal domain of Cbl complexed to its binding site on ZAP-70 kinase.
Nature, 398, 1999
|
|
1FMK
 
 | |
1IJQ
 
 | | Crystal Structure of the LDL Receptor YWTD-EGF Domain Pair | | Descriptor: | LOW-DENSITY LIPOPROTEIN RECEPTOR | | Authors: | Jeon, H, Meng, W, Takagi, J, Eck, M.J, Springer, T.A, Blacklow, S.C. | | Deposit date: | 2001-04-27 | | Release date: | 2001-05-23 | | Last modified: | 2018-04-04 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Implications for familial hypercholesterolemia from the structure of the LDL receptor YWTD-EGF domain pair.
Nat.Struct.Biol., 8, 2001
|
|
1OX9
 
 | | Crystal structure of SspB-ssrA complex | | Descriptor: | Stringent starvation protein B, ssrA | | Authors: | Song, H.K, Eck, M.J. | | Deposit date: | 2003-04-01 | | Release date: | 2003-08-26 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural basis of degradation signal recognition by SspB, a specificity-enhancing factor for the ClpXP proteolytic machine
Mol.Cell, 12, 2003
|
|
7JXL
 
 | | EGFR kinase (T790M/V948R) in complex with AZ5104 | | Descriptor: | Epidermal growth factor receptor, N-(2-{[2-(dimethylamino)ethyl](methyl)amino}-5-{[4-(1H-indol-3-yl)pyrimidin-2-yl]amino}-4-methoxyphenyl)propanamide | | Authors: | Beyett, T.S, Eck, M.J. | | Deposit date: | 2020-08-27 | | Release date: | 2021-09-08 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Molecular basis for cooperative binding and synergy of ATP-site and allosteric EGFR inhibitors
Nat Commun, 13, 2022
|
|
7K1I
 
 | | EGFR kinase (L858R/V948R) in complex with allosteric inhibitor JBJ-09-063 | | Descriptor: | (2R)-2-(5-fluoro-2-hydroxyphenyl)-2-{6-[4-(1-methylpiperidin-4-yl)phenyl]-1-oxo-1,3-dihydro-2H-isoindol-2-yl}-N-(1,3-thiazol-2-yl)acetamide, Epidermal growth factor receptor, MAGNESIUM ION, ... | | Authors: | Beyett, T.S, Eck, M.J. | | Deposit date: | 2020-09-07 | | Release date: | 2021-09-15 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.202 Å) | | Cite: | Molecular basis for cooperative binding and synergy of ATP-site and allosteric EGFR inhibitors
Nat Commun, 13, 2022
|
|
7JXI
 
 | | EGFR kinase (T790M/V948R) in complex with PF-06747775 | | Descriptor: | Epidermal growth factor receptor, N-[(3R,4R)-4-fluoro-1-{6-[(3-methoxy-1-methyl-1H-pyrazol-4-yl)amino]-9-methyl-9H-purin-2-yl}pyrrolidin-3-yl]propanamide | | Authors: | Beyett, T.S, Eck, M.J. | | Deposit date: | 2020-08-27 | | Release date: | 2021-09-08 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Molecular basis for cooperative binding and synergy of ATP-site and allosteric EGFR inhibitors
Nat Commun, 13, 2022
|
|
1I1Q
 
 | |
6Q0T
 
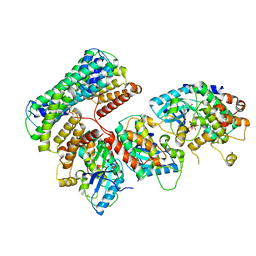 | | Structure of a MAPK pathway complex | | Descriptor: | 14-3-3 protein zeta, 5-[(2-fluoro-4-iodophenyl)amino]-N-(2-hydroxyethoxy)imidazo[1,5-a]pyridine-6-carboxamide, Dual specificity mitogen-activated protein kinase kinase 1, ... | | Authors: | Park, E, Rawson, S, Jeon, H, Eck, M.J. | | Deposit date: | 2019-08-02 | | Release date: | 2019-10-09 | | Last modified: | 2020-04-22 | | Method: | ELECTRON MICROSCOPY (5.7 Å) | | Cite: | Architecture of autoinhibited and active BRAF-MEK1-14-3-3 complexes.
Nature, 575, 2019
|
|
1NTV
 
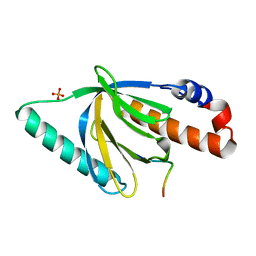 | | Crystal Structure of the Disabled-1 (Dab1) PTB domain-ApoER2 peptide complex | | Descriptor: | Apolipoprotein E Receptor-2 peptide, Disabled homolog 1, PHOSPHATE ION | | Authors: | Stolt, P.C, Jeon, H, Song, H.K, Herz, J, Eck, M.J, Blacklow, S.C. | | Deposit date: | 2003-01-30 | | Release date: | 2003-04-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Origins of Peptide Selectivity and Phosphoinositide Binding Revealed by Structures of Disabled-1 PTB Domain Complexes
Structure, 11, 2003
|
|
1OX8
 
 | | Crystal structure of SspB | | Descriptor: | Stringent starvation protein B | | Authors: | Song, H.K, Eck, M.J. | | Deposit date: | 2003-04-01 | | Release date: | 2003-08-26 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis of degradation signal recognition by SspB, a specificity-enhancing factor for the ClpXP proteolytic machine
Mol.Cell, 12, 2003
|
|
1Q68
 
 | | Solution structure of T-cell surface glycoprotein CD4 and Proto-oncogene tyrosine-protein kinase LCK fragments | | Descriptor: | Proto-oncogene tyrosine-protein kinase LCK, T-cell surface glycoprotein CD4, ZINC ION | | Authors: | Kim, P.W, Sun, Z.Y, Blacklow, S.C, Wagner, G, Eck, M.J. | | Deposit date: | 2003-08-12 | | Release date: | 2003-11-25 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | A zinc clasp structure tethers Lck to T cell coreceptors CD4 and CD8.
Science, 301, 2003
|
|
1P4Q
 
 | | Solution structure of the CITED2 transactivation domain in complex with the p300 CH1 domain | | Descriptor: | Cbp/p300-interacting transactivator 2, E1A-associated protein p300, ZINC ION | | Authors: | Freedman, S.J, Sun, Z.-Y.J, Kung, A.L, France, D.S, Wagner, G, Eck, M.J. | | Deposit date: | 2003-04-23 | | Release date: | 2003-07-01 | | Last modified: | 2017-02-01 | | Method: | SOLUTION NMR | | Cite: | Structural basis for negative regulation of hypoxia-inducible factor-1alpha by CITED2.
Nat.Struct.Biol., 10, 2003
|
|
1Q69
 
 | | Solution structure of T-cell surface glycoprotein CD8 alpha chain and Proto-oncogene tyrosine-protein kinase LCK fragments | | Descriptor: | Proto-oncogene tyrosine-protein kinase LCK, T-cell surface glycoprotein CD8 alpha chain, ZINC ION | | Authors: | Kim, P.W, Sun, Z.Y, Blacklow, S.C, Wagner, G, Eck, M.J. | | Deposit date: | 2003-08-12 | | Release date: | 2003-11-25 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | A zinc clasp structure tethers Lck to T cell coreceptors CD4 and CD8.
Science, 301, 2003
|
|
3ZDT
 
 | | Crystal structure of basic patch mutant FAK FERM domain FAK31- 405 K216A, K218A, R221A, K222A | | Descriptor: | FOCAL ADHESION KINASE 1 | | Authors: | Goni, G.M, Epifano, C, Boskovic, J, Camacho-Artacho, M, Zhou, J, Martin, M.T, Eck, M.J, Kremer, L, Graeter, F, Gervasio, F.L, Perez-Moreno, M, Lietha, D. | | Deposit date: | 2012-11-30 | | Release date: | 2012-12-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Phosphatidylinositol 4,5-Bisphosphate Triggers Activation of Focal Adhesion Kinase by Inducing Clustering and Conformational Changes.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4CYE
 
 | | Crystal structure of avian FAK FERM domain FAK31-405 at 3.2A | | Descriptor: | FOCAL ADHESION KINASE 1 | | Authors: | Goni, G.M, Epifano, C, Boskovic, J, Camacho-Artacho, M, Zhou, J, Martin, M.T, Eck, M.J, Kremer, L, Graeter, F, Gervasio, F.L, Perez-Moreno, M, Lietha, D. | | Deposit date: | 2014-04-10 | | Release date: | 2014-04-23 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Phosphatidylinositol 4,5-Bisphosphate Triggers Activation of Focal Adhesion Kinase by Inducing Clustering and Conformational Changes.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
