6AVB
 
 | | CryoEM structure of Mical Oxidized Actin (Class 1) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin, alpha skeletal muscle | | Authors: | Grintsevich, E.E, Ge, P, Sawaya, M.R, Yesilyurt, H.G, Terman, J.R, Zhou, Z.H, Reisler, E. | | Deposit date: | 2017-09-01 | | Release date: | 2018-01-17 | | Last modified: | 2019-11-06 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Catastrophic disassembly of actin filaments via Mical-mediated oxidation.
Nat Commun, 8, 2017
|
|
8GK3
 
 | | Cytochrome P450 3A7 in complex with Dehydroepiandrosterone sulfate | | Descriptor: | 17-oxoandrost-5-en-3beta-yl hydrogen sulfate, Cytochrome P450 3A7, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Liu, J, Scott, E.E. | | Deposit date: | 2023-03-16 | | Release date: | 2023-07-26 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Human cytochrome P450 3A7 binding four copies of its native substrate dehydroepiandrosterone 3-sulfate.
J.Biol.Chem., 299, 2023
|
|
6AV9
 
 | | CryoEM structure of Mical Oxidized Actin (Class 1) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin, alpha skeletal muscle | | Authors: | Grintsevich, E.E, Ge, P, Sawaya, M.R, Yesilyurt, H.G, Terman, J.R, Zhou, Z.H, Reisler, E. | | Deposit date: | 2017-09-01 | | Release date: | 2018-01-17 | | Last modified: | 2019-11-27 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Catastrophic disassembly of actin filaments via Mical-mediated oxidation.
Nat Commun, 8, 2017
|
|
8G2D
 
 | | Crystal structure of the wild-type Thermus thermophilus 70S ribosome in complex with tylosin, mRNA, deacylated A- and E-site tRNAphe, and deacylated P-site tRNAmet at 2.70A resolution | | Descriptor: | 16S Ribosomal RNA, 23S Ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Aleksandrova, E.V, Wu, K.J.Y, Tresco, B.I.C, Syroegin, E.A, Killeavy, E.E, Balasanyants, S.M, Svetlov, M.S, Gregory, S.T, Atkinson, G.C, Myers, A.G, Polikanov, Y.S. | | Deposit date: | 2023-02-03 | | Release date: | 2023-12-27 | | Last modified: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis of Cfr-mediated antimicrobial resistance and mechanisms to evade it.
Nat.Chem.Biol., 20, 2024
|
|
8G6D
 
 | |
8G29
 
 | | Crystal structure of the A2503-C2,C8-dimethylated Thermus thermophilus 70S ribosome in complex with mRNA, aminoacylated A-site Phe-tRNAphe, aminoacylated P-site fMet-tRNAmet, and deacylated E-site tRNAphe at 2.55A resolution | | Descriptor: | 16S Ribosomal RNA, 23S Ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Aleksandrova, E.V, Wu, K.J.Y, Tresco, B.I.C, Syroegin, E.A, Killeavy, E.E, Balasanyants, S.M, Svetlov, M.S, Gregory, S.T, Atkinson, G.C, Myers, A.G, Polikanov, Y.S. | | Deposit date: | 2023-02-03 | | Release date: | 2023-12-27 | | Last modified: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structural basis of Cfr-mediated antimicrobial resistance and mechanisms to evade it.
Nat.Chem.Biol., 20, 2024
|
|
8FDA
 
 | | Human Cytochrome P450 17A1 in complex with steroidal isonitrile inhibitor | | Descriptor: | PROTOPORPHYRIN IX CONTAINING FE, Steroid 17-alpha-hydroxylase/17,20 lyase, [(1~{R})-1-[(3~{S},5~{S},8~{R},9~{S},10~{S},13~{S},17~{R})-3-methanoyloxy-10,13-dimethyl-1,2,3,4,5,6,7,8,9,11,12,13,14,15,16,17-hexadecahydrocyclopenta[a]phenanthren-17-yl]ethyl]-methylidyne-azanium | | Authors: | Richard, A.M, Scott, E.E. | | Deposit date: | 2022-12-02 | | Release date: | 2023-08-23 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Selective steroidogenic cytochrome P450 haem iron ligation by steroid-derived isonitriles.
Commun Chem, 6, 2023
|
|
8G2C
 
 | | Crystal structure of the A2503-C2,C8-dimethylated Thermus thermophilus 70S ribosome in complex with tylosin, mRNA, aminoacylated A-site Phe-tRNAphe, aminoacylated P-site fMet-tRNAmet, and deacylated E-site tRNAphe at 2.65A resolution | | Descriptor: | 16S Ribosomal RNA, 23S Ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Aleksandrova, E.V, Wu, K.J.Y, Tresco, B.I.C, Syroegin, E.A, Killeavy, E.E, Balasanyants, S.M, Svetlov, M.S, Gregory, S.T, Atkinson, G.C, Myers, A.G, Polikanov, Y.S. | | Deposit date: | 2023-02-03 | | Release date: | 2023-12-27 | | Last modified: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structural basis of Cfr-mediated antimicrobial resistance and mechanisms to evade it.
Nat.Chem.Biol., 20, 2024
|
|
8G2B
 
 | | Crystal structure of the A2503-C2,C8-dimethylated Thermus thermophilus 70S ribosome in complex with iboxamycin, mRNA, deacylated A- and E-site tRNAphe, and aminoacylated P-site fMet-tRNAmet at 2.55A resolution | | Descriptor: | 16S Ribosomal RNA, 23S Ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Aleksandrova, E.V, Wu, K.J.Y, Tresco, B.I.C, Syroegin, E.A, Killeavy, E.E, Balasanyants, S.M, Svetlov, M.S, Gregory, S.T, Atkinson, G.C, Myers, A.G, Polikanov, Y.S. | | Deposit date: | 2023-02-03 | | Release date: | 2023-12-27 | | Last modified: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structural basis of Cfr-mediated antimicrobial resistance and mechanisms to evade it.
Nat.Chem.Biol., 20, 2024
|
|
8G2A
 
 | | Crystal structure of the A2503-C2,C8-dimethylated Thermus thermophilus 70S ribosome in complex with mRNA, aminoacylated A-site Phe-tRNAphe, peptidyl P-site fMTHSMRC-tRNAmet, and deacylated E-site tRNAphe at 2.45A resolution | | Descriptor: | 16S Ribosomal RNA, 23S Ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Aleksandrova, E.V, Wu, K.J.Y, Tresco, B.I.C, Syroegin, E.A, Killeavy, E.E, Balasanyants, S.M, Svetlov, M.S, Gregory, S.T, Atkinson, G.C, Myers, A.G, Polikanov, Y.S. | | Deposit date: | 2023-02-03 | | Release date: | 2023-12-27 | | Last modified: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structural basis of Cfr-mediated antimicrobial resistance and mechanisms to evade it.
Nat.Chem.Biol., 20, 2024
|
|
8HRG
 
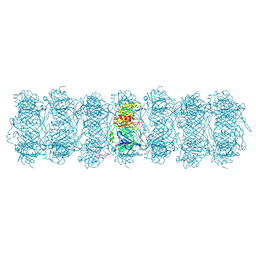 | | Tail tube of DT57C bacteriophage in the full state | | Descriptor: | Tail tube protein | | Authors: | Ayala, R, Moiseenko, A.V, Chen, T.H, Kulikov, E.E, Golomidova, A.K, Orekhov, P.S, Street, M.A, Sokolova, O.S, Letarov, A.V, Wolf, M. | | Deposit date: | 2022-12-15 | | Release date: | 2023-12-13 | | Last modified: | 2023-12-27 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Nearly complete structure of bacteriophage DT57C reveals architecture of head-to-tail interface and lateral tail fibers.
Nat Commun, 14, 2023
|
|
8HQK
 
 | | Capsid of DT57C bacteriophage in the empty state | | Descriptor: | Major head protein | | Authors: | Ayala, R, Moiseenko, A.V, Kulikov, E.E, Golomidova, A.K, Orekhov, P.S, Street, M.A, Sokolova, O.S, Letarov, A.V, Wolf, M. | | Deposit date: | 2022-12-13 | | Release date: | 2023-12-13 | | Last modified: | 2023-12-27 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Nearly complete structure of bacteriophage DT57C reveals architecture of head-to-tail interface and lateral tail fibers.
Nat Commun, 14, 2023
|
|
8HRE
 
 | | Straight fiber of DT57C bacteriophage in the full state | | Descriptor: | Central straight fiber | | Authors: | Ayala, R, Moiseenko, A.V, Chen, T.H, Kulikov, E.E, Golomidova, A.K, Orekhov, P.S, Street, M.A, Sokolova, O.S, Letarov, A.V, Wolf, M. | | Deposit date: | 2022-12-15 | | Release date: | 2023-12-13 | | Last modified: | 2023-12-27 | | Method: | ELECTRON MICROSCOPY (5.5 Å) | | Cite: | Nearly complete structure of bacteriophage DT57C reveals architecture of head-to-tail interface and lateral tail fibers.
Nat Commun, 14, 2023
|
|
8HO3
 
 | | Capsid of DT57C bacteriophage in the full state | | Descriptor: | Major head protein | | Authors: | Ayala, R, Moiseenko, A.V, Chen, T.H, Kulikov, E.E, Golomidova, A.K, Orekhov, P.S, Street, M.A, Sokolova, O.S, Letarov, A.V, Wolf, M. | | Deposit date: | 2022-12-09 | | Release date: | 2023-12-13 | | Last modified: | 2023-12-27 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Nearly complete structure of bacteriophage DT57C reveals architecture of head-to-tail interface and lateral tail fibers.
Nat Commun, 14, 2023
|
|
6CHI
 
 | | Human Cytochrome P450 17A1 in complex with inhibitor: abiraterone C6 amide | | Descriptor: | 3-hydroxy-17-(3-pyridyl)-androst-5,16-dien-6-amide, PROTOPORPHYRIN IX CONTAINING FE, Steroid 17-alpha-hydroxylase/17,20 lyase | | Authors: | Scott, E.E. | | Deposit date: | 2018-02-22 | | Release date: | 2018-06-06 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.698 Å) | | Cite: | Structure-Based Design of Inhibitors with Improved Selectivity for Steroidogenic Cytochrome P450 17A1 over Cytochrome P450 21A2.
J. Med. Chem., 61, 2018
|
|
8HQZ
 
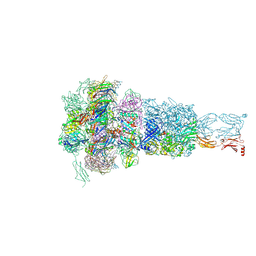 | | Baseplate of DT57C bacteriophage in the full state | | Descriptor: | Baseplate hub protein, Distal tail protein, L-shaped tail fiber assembly, ... | | Authors: | Ayala, R, Moiseenko, A.V, Chen, T.H, Kulikov, E.E, Golomidova, A.K, Orekhov, P.S, Street, M.A, Sokolova, O.S, Letarov, A.V, Wolf, M. | | Deposit date: | 2022-12-14 | | Release date: | 2023-12-13 | | Last modified: | 2023-12-27 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Nearly complete structure of bacteriophage DT57C reveals architecture of head-to-tail interface and lateral tail fibers.
Nat Commun, 14, 2023
|
|
8HQO
 
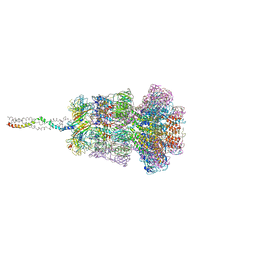 | | Neck of DT57C bacteriophage in the full state | | Descriptor: | Head completion protein, Portal protein, Tail terminator protein, ... | | Authors: | Ayala, R, Moiseenko, A.V, Chen, T.H, Kulikov, E.E, Golomidova, A.K, Orekhov, P.S, Street, M.A, Sokolova, O.S, Letarov, A.V, Wolf, M. | | Deposit date: | 2022-12-13 | | Release date: | 2023-12-13 | | Last modified: | 2023-12-27 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Nearly complete structure of bacteriophage DT57C reveals architecture of head-to-tail interface and lateral tail fibers.
Nat Commun, 14, 2023
|
|
6BM8
 
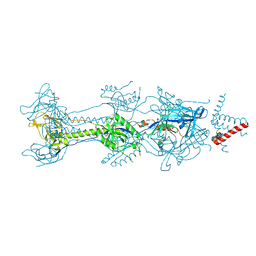 | | Crystal structure of glycoprotein B from Herpes Simplex Virus type I | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope glycoprotein B, ... | | Authors: | Cooper, R.S, Heldwein, E.E. | | Deposit date: | 2017-11-13 | | Release date: | 2018-05-16 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (4.1 Å) | | Cite: | Structural basis for membrane anchoring and fusion regulation of the herpes simplex virus fusogen gB.
Nat. Struct. Mol. Biol., 25, 2018
|
|
6CIR
 
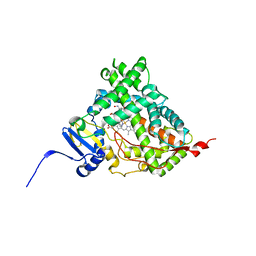 | | Human Cytochrome P450 17A1 in complex with inhibitor: abiraterone C6 oxime | | Descriptor: | 6-oxime-17-(3-pyridyl)-androst-16-en-3-ol, PROTOPORPHYRIN IX CONTAINING FE, Steroid 17-alpha-hydroxylase/17,20 lyase | | Authors: | Scott, E.E, Fehl, C. | | Deposit date: | 2018-02-25 | | Release date: | 2018-06-06 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.648 Å) | | Cite: | Structure-Based Design of Inhibitors with Improved Selectivity for Steroidogenic Cytochrome P450 17A1 over Cytochrome P450 21A2.
J. Med. Chem., 61, 2018
|
|
4MIK
 
 | | Crystal Structure of hPNMT in Complex with bisubstrate inhibitor (2R,3R,4S,5S)-2-(6-amino-9H-purin-9-yl)-5-(((2-(((7-nitro-1,2,3,4-tetrahydroisoquinolin-3-yl)methyl)amino)ethyl)thio)methyl)tetrahydrofuran-3,4-diol | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 9-{5-S-[2-({[(3S)-7-nitro-1,2,3,4-tetrahydroisoquinolin-3-yl]methyl}amino)ethyl]-5-thio-alpha-L-lyxofuranosyl}-9H-purin-6-amine, Phenylethanolamine N-methyltransferase | | Authors: | Bart, A.G, Scott, E.E. | | Deposit date: | 2013-08-31 | | Release date: | 2014-09-10 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (1.9506 Å) | | Cite: | To be published
To be Published
|
|
4OGW
 
 | | Structure of a human CD38 mutant complexed with NMN | | Descriptor: | ADP-ribosyl cyclase 1, BETA-NICOTINAMIDE RIBOSE MONOPHOSPHATE | | Authors: | Shewchuk, L.M, Preugschat, F, Carter, L.H, Boros, E.E, Moyer, M.B, Stewart, E.L, Porter, D.J.T. | | Deposit date: | 2014-01-16 | | Release date: | 2014-10-08 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | A pre-steady state and steady state kinetic analysis of the N-ribosyl hydrolase activity of hCD157.
Arch.Biochem.Biophys., 564C, 2014
|
|
4NKX
 
 | |
4NKZ
 
 | | Human steroidogenic cytochrome P450 17A1 mutant A105L with substrate 17alpha-hydroxypregnenolone | | Descriptor: | (3alpha,8alpha)-3,17-dihydroxypregn-5-en-20-one, PROTOPORPHYRIN IX CONTAINING FE, Steroid 17-alpha-hydroxylase/17,20 lyase | | Authors: | Scott, E.E, Petrunak, E.M. | | Deposit date: | 2013-11-13 | | Release date: | 2014-10-22 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.003 Å) | | Cite: | Structures of Human Steroidogenic Cytochrome P450 17A1 with Substrates.
J.Biol.Chem., 289, 2014
|
|
4MQ4
 
 | | Crystal Structure of hPNMT in Complex with bisubstrate inhibitor N-(3-((((2S,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-3,4-dihydroxytetrahydrofuran-2-yl)methyl)thio)propyl)-1,2,3,4-tetrahydroisoquinoline-3-carboxamide | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 5'-S-(3-{[(3R)-1,2,3,4-tetrahydroisoquinolin-3-ylcarbonyl]amino}propyl)-5'-thioadenosine, Phenylethanolamine N-methyltransferase | | Authors: | Bart, A.G, Scott, E.E. | | Deposit date: | 2013-09-15 | | Release date: | 2014-10-15 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (2.2042 Å) | | Cite: | To be Published
To be Published
|
|
1WS0
 
 | |
