2PUY
 
 | |
7MGA
 
 | | Concanavalin A bound to a DNA glycoconjugate, Man-AAATTT | | Descriptor: | 3-{[2-(2-hydroxyethoxy)ethyl]amino}-4-[(6-hydroxyhexyl)amino]cyclobut-3-ene-1,2-dione, CALCIUM ION, Concanavalin-A, ... | | Authors: | Partridge, B.E, Winegar, P.H, Mirkin, C.A. | | Deposit date: | 2021-04-12 | | Release date: | 2021-06-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Redefining Protein Interfaces within Protein Single Crystals with DNA.
J.Am.Chem.Soc., 143, 2021
|
|
6TOW
 
 | |
7MJS
 
 | | Single-Particle Cryo-EM Structure of Major Facilitator Superfamily Domain containing 2A in complex with LPC-18:3 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2AG3 Fab heavy chain, 2AG3 Fab light chain, ... | | Authors: | Cater, R.J, Chua, G.L, Erramilli, S.K, Keener, J.E, Choy, B.C, Tokarz, P, Chin, C.F, Quek, D.Q.Y, Kloss, B, Pepe, J.G, Parisi, G, Wong, B.H, Clarke, O.B, Marty, M.T, Kossiakoff, A.A, Khelashvili, G, Silver, D.L, Mancia, F. | | Deposit date: | 2021-04-20 | | Release date: | 2021-06-16 | | Last modified: | 2021-07-28 | | Method: | ELECTRON MICROSCOPY (3.03 Å) | | Cite: | Structural basis of omega-3 fatty acid transport across the blood-brain barrier.
Nature, 595, 2021
|
|
7MG5
 
 | | Concanavalin A bound to a DNA glycoconjugate, Man-ATAT | | Descriptor: | 3-{[2-(2-hydroxyethoxy)ethyl]amino}-4-[(6-hydroxyhexyl)amino]cyclobut-3-ene-1,2-dione, CALCIUM ION, Concanavalin-A, ... | | Authors: | Partridge, B.E, Winegar, P.H, Mirkin, C.A. | | Deposit date: | 2021-04-12 | | Release date: | 2021-06-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Redefining Protein Interfaces within Protein Single Crystals with DNA.
J.Am.Chem.Soc., 143, 2021
|
|
7MG8
 
 | | Concanavalin A bound to a DNA glycoconjugate, Man-CGCG | | Descriptor: | 3-{[2-(2-hydroxyethoxy)ethyl]amino}-4-[(6-hydroxyhexyl)amino]cyclobut-3-ene-1,2-dione, CALCIUM ION, Concanavalin-A, ... | | Authors: | Partridge, B.E, Winegar, P.H, Mirkin, C.A. | | Deposit date: | 2021-04-12 | | Release date: | 2021-06-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Redefining Protein Interfaces within Protein Single Crystals with DNA.
J.Am.Chem.Soc., 143, 2021
|
|
4NJA
 
 | | Crystal structure of Fab 6C8 in complex with MPTS | | Descriptor: | 6C8 heavy chain, 6C8 light chain, 8-methoxypyrene-1,3,6-trisulfonic acid, ... | | Authors: | Stanfield, R.L, Romesberg, F.E, Zimmermann, J, Wilson, I.A. | | Deposit date: | 2013-11-08 | | Release date: | 2014-11-12 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.204 Å) | | Cite: | Adaptive Mutations Alter Antibody Structure and Dynamics during Affinity Maturation.
Biochemistry, 54, 2015
|
|
7MGB
 
 | | Concanavalin A bound to a DNA glycoconjugate, A(Man-T)AT | | Descriptor: | CALCIUM ION, Concanavalin-A, MANGANESE (II) ION, ... | | Authors: | Partridge, B.E, Winegar, P.H, Mirkin, C.A. | | Deposit date: | 2021-04-12 | | Release date: | 2021-06-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Redefining Protein Interfaces within Protein Single Crystals with DNA.
J.Am.Chem.Soc., 143, 2021
|
|
7MG6
 
 | | Concanavalin A bound to a DNA glycoconjugate, Man-AGCT | | Descriptor: | 3-{[2-(2-hydroxyethoxy)ethyl]amino}-4-[(6-hydroxyhexyl)amino]cyclobut-3-ene-1,2-dione, CALCIUM ION, Concanavalin-A, ... | | Authors: | Partridge, B.E, Winegar, P.H, Mirkin, C.A. | | Deposit date: | 2021-04-12 | | Release date: | 2021-06-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Redefining Protein Interfaces within Protein Single Crystals with DNA.
J.Am.Chem.Soc., 143, 2021
|
|
7MG9
 
 | | Concanavalin A bound to DNA glycoconjugates, Man-TTTT and Man-AAAA | | Descriptor: | 3-{[2-(2-hydroxyethoxy)ethyl]amino}-4-[(6-hydroxyhexyl)amino]cyclobut-3-ene-1,2-dione, CALCIUM ION, Concanavalin-A, ... | | Authors: | Partridge, B.E, Winegar, P.H, Mirkin, C.A. | | Deposit date: | 2021-04-12 | | Release date: | 2021-06-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Redefining Protein Interfaces within Protein Single Crystals with DNA.
J.Am.Chem.Soc., 143, 2021
|
|
4NKE
 
 | | The effects of Lysine 200 and Phenylalanine 239 Farnesyl Pyrophosphate Synthase (FPPS) mutations on the catalytic activity, crystal structure and inhibition by nitrogen containing bisphosphonates | | Descriptor: | 1,2-ETHANEDIOL, 1-HYDROXY-2-(3-PYRIDINYL)ETHYLIDENE BIS-PHOSPHONIC ACID, 3-METHYLBUT-3-ENYL TRIHYDROGEN DIPHOSPHATE, ... | | Authors: | Tsoumpra, M.K, Barnett, B.L, Muniz, J.R.C, Walter, R.L, Ebetino, F.H, von Delft, F, Russell, R.G.G, Oppermann, U, Dunford, J.E. | | Deposit date: | 2013-11-12 | | Release date: | 2014-11-19 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | The effects of Lysine 200 and Phenylalanine 239 Farnesyl Pyrophosphate Synthase (FPPS) mutations on the catalytic activity, crystal structure and inhibition by nitrogen containing bisphosphonates
To be Published
|
|
6DW1
 
 | | Cryo-EM structure of the benzodiazepine-sensitive alpha1beta1gamma2S tri-heteromeric GABAA receptor in complex with GABA (ECD map) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, GAMMA-AMINO-BUTANOIC ACID, Gamma-aminobutyric acid receptor subunit alpha-1,Gamma-aminobutyric acid receptor subunit alpha-1, ... | | Authors: | Phulera, S, Zhu, H, Yu, J, Yoshioka, C, Gouaux, E. | | Deposit date: | 2018-06-26 | | Release date: | 2018-08-08 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Cryo-EM structure of the benzodiazepine-sensitive alpha 1 beta 1 gamma 2S tri-heteromeric GABAAreceptor in complex with GABA.
Elife, 7, 2018
|
|
6TR6
 
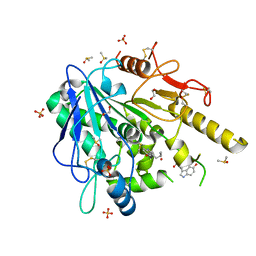 | | N-acetylserotonin-Notum complex | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, DIMETHYL SULFOXIDE, ... | | Authors: | Zhao, Y, Jones, E.Y. | | Deposit date: | 2019-12-17 | | Release date: | 2020-01-08 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Structural characterization of melatonin as an inhibitor of the Wnt deacylase Notum.
J. Pineal Res., 68, 2020
|
|
4NKQ
 
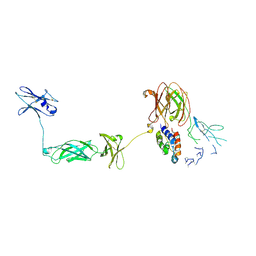 | | Structure of a Cytokine Receptor Complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Cytokine receptor common subunit beta, Granulocyte-macrophage colony-stimulating factor, ... | | Authors: | Parker, M.W, Broughton, S.E. | | Deposit date: | 2013-11-13 | | Release date: | 2015-09-23 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3.301 Å) | | Cite: | Conformational Changes in the GM-CSF Receptor Suggest a Molecular Mechanism for Affinity Conversion and Receptor Signaling.
Structure, 24, 2016
|
|
7MGC
 
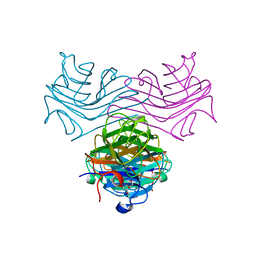 | | Concanavalin A bound to a DNA glycoconjugate, G(Man-T)AC | | Descriptor: | CALCIUM ION, Concanavalin-A, MANGANESE (II) ION, ... | | Authors: | Partridge, B.E, Winegar, P.H, Mirkin, C.A. | | Deposit date: | 2021-04-12 | | Release date: | 2021-06-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.92 Å) | | Cite: | Redefining Protein Interfaces within Protein Single Crystals with DNA.
J.Am.Chem.Soc., 143, 2021
|
|
7MGS
 
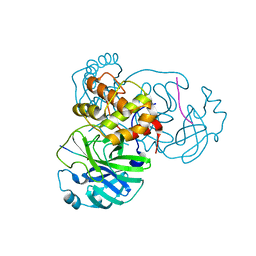 | |
2Q3G
 
 | | Structure of the PDZ domain of human PDLIM7 bound to a C-terminal extension from human beta-tropomyosin | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, PDZ and LIM domain protein 7 | | Authors: | Gileadi, C, Papagrigoriou, E, Elkins, J, Burgess-Brown, N, Salah, E, Gileadi, O, Umeano, C, Bunkoczi, G, von Delft, F, Uppenberg, J, Pike, A.C.W, Arrowsmith, C.H, Edwards, A, Weigelt, J, Sundstrom, M, Doyle, D.A, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-05-30 | | Release date: | 2007-06-19 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.11 Å) | | Cite: | Unusual binding interactions in PDZ domain crystal structures help explain binding mechanisms
Protein Sci., 19, 2010
|
|
2Q5I
 
 | | Crystal structure of apo S581L Glycyl-tRNA synthetase mutant | | Descriptor: | Glycyl-tRNA synthetase, SULFATE ION | | Authors: | Cader, M.Z, Ren, J, James, P.A, Bird, L.E, Talbot, K, Stammers, D.K, Oxford Protein Production Facility (OPPF) | | Deposit date: | 2007-06-01 | | Release date: | 2007-06-19 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of human wildtype and S581L-mutant glycyl-tRNA synthetase, an enzyme underlying distal spinal muscular atrophy.
Febs Lett., 581, 2007
|
|
4NT0
 
 | | Crystal structure of orotidine 5'-monophosphate decarboxylase from Methanobacterium thermoautotrophicum complexed with 3-deazauridine 5'-monophosphate | | Descriptor: | 4-hydroxy-1-(5-O-phosphono-beta-D-ribofuranosyl)pyridin-2(1H)-one, CALCIUM ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Fedorov, A.A, Fedorov, E.V, Iiams, V, Gerlt, J.A, Almo, S.C. | | Deposit date: | 2013-11-29 | | Release date: | 2013-12-18 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.769 Å) | | Cite: | Crystal structure of orotidine 5'-monophosphate decarboxylase from Methanobacterium thermoautotrophicum complexed with 3-deazauridine 5'-monophosphate
To be Published
|
|
4NTE
 
 | | Crystal structure of DepH | | Descriptor: | CHLORIDE ION, DepH, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Scharf, D.H, Groll, M, Habel, A, Heinekamp, T, Hertweck, C, Brakhage, A.A, Huber, E.M. | | Deposit date: | 2013-12-02 | | Release date: | 2014-03-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Flavoenzyme-Catalyzed Formation of Disulfide Bonds in Natural Products
Angew.Chem.Int.Ed.Engl., 53, 2014
|
|
7M8X
 
 | | CRYSTAL STRUCTURE OF THE SARS-COV-2(2019-NCOV) MAIN PROTEASE IN COMPLEX WITH COMPOUND 6 | | Descriptor: | 2-{3-[3-chloro-5-(2-methoxyethoxy)phenyl]-2-oxo-2H-[1,3'-bipyridin]-5-yl}benzonitrile, 3C-like proteinase | | Authors: | Deshmukh, M.G, Ippolito, J.A, Stone, E.A, Jorgensen, W.L, Anderson, K.S. | | Deposit date: | 2021-03-30 | | Release date: | 2021-06-30 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Structure-guided design of a perampanel-derived pharmacophore targeting the SARS-CoV-2 main protease.
Structure, 29, 2021
|
|
2PLJ
 
 | |
7MBR
 
 | | Cryo-EM structure of zebrafish TRPM5 in the presence of 6 uM calcium (apo state) | | Descriptor: | (25R)-14beta,17beta-spirost-5-en-3beta-ol, (2R)-2-(hydroxymethyl)-4-{[(25R)-10alpha,14beta,17beta-spirost-5-en-3beta-yl]oxy}butyl 4-O-alpha-D-glucopyranosyl-beta-D-glucopyranoside, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Ruan, Z, Lu, W, Du, J, Haley, E. | | Deposit date: | 2021-04-01 | | Release date: | 2021-07-07 | | Last modified: | 2021-07-28 | | Method: | ELECTRON MICROSCOPY | | Cite: | Structures of the TRPM5 channel elucidate mechanisms of activation and inhibition.
Nat.Struct.Mol.Biol., 28, 2021
|
|
4NX7
 
 | | single cryogenic temperature model of DHFR | | Descriptor: | BETA-MERCAPTOETHANOL, Dihydrofolate reductase, FOLIC ACID, ... | | Authors: | Fenwick, R.B, van den Bedem, H, Fraser, J.S, Wright, P.E. | | Deposit date: | 2013-12-08 | | Release date: | 2014-01-15 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Integrated description of protein dynamics from room-temperature X-ray crystallography and NMR.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
7MBS
 
 | | Cryo-EM structure of zebrafish TRPM5 in the presence of 6 uM calcium (open state) | | Descriptor: | (25R)-14beta,17beta-spirost-5-en-3beta-ol, (2R)-2-(hydroxymethyl)-4-{[(25R)-10alpha,14beta,17beta-spirost-5-en-3beta-yl]oxy}butyl 4-O-alpha-D-glucopyranosyl-beta-D-glucopyranoside, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Ruan, Z, Lu, W, Du, J, Haley, E. | | Deposit date: | 2021-04-01 | | Release date: | 2021-07-07 | | Last modified: | 2021-07-28 | | Method: | ELECTRON MICROSCOPY | | Cite: | Structures of the TRPM5 channel elucidate mechanisms of activation and inhibition.
Nat.Struct.Mol.Biol., 28, 2021
|
|
