6S0S
 
 | | The crystal structure of kanamycin B dioxygenase (KanJ) from Streptomyces kanamyceticus in complex with nickel, ribostamycin B and 2-oxoglutarate | | Descriptor: | 2-OXOGLUTARIC ACID, CHLORIDE ION, Kanamycin B dioxygenase, ... | | Authors: | Mrugala, B, Porebski, P.J, Niedzialkowska, E, Minor, W, Borowski, T. | | Deposit date: | 2019-06-18 | | Release date: | 2020-07-08 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | A study on the structure, mechanism, and biochemistry of kanamycin B dioxygenase (KanJ)-an enzyme with a broad range of substrates.
Febs J., 288, 2021
|
|
4F6M
 
 | | Crystal structure of Kaiso zinc finger DNA binding domain in complex with Kaiso binding site DNA | | Descriptor: | DNA (5'-D(*CP*GP*TP*TP*AP*TP*TP*GP*GP*CP*AP*GP*GP*AP*AP*GP*CP*AP*C)-3'), DNA (5'-D(*GP*TP*GP*CP*TP*TP*CP*CP*TP*GP*CP*CP*AP*AP*TP*AP*AP*CP*G)-3'), Transcriptional regulator Kaiso, ... | | Authors: | Buck-Koehntop, B.A, Stanfield, R.L, Ekiert, D.C, Martinez-Yamout, M.A, Dyson, H.J, Wilson, I.A, Wright, P.E. | | Deposit date: | 2012-05-15 | | Release date: | 2012-09-05 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Molecular basis for recognition of methylated and specific DNA sequences by the zinc finger protein Kaiso.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
6CYX
 
 | |
8VFU
 
 | |
3NMH
 
 | | Crystal structure of the abscisic receptor PYL2 in complex with pyrabactin | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, 4-bromo-N-(pyridin-2-ylmethyl)naphthalene-1-sulfonamide, Abscisic acid receptor PYL2 | | Authors: | Zhou, X.E, Melcher, K, Ng, L.-M, Soon, F.-F, Xu, Y, Suino-Powell, K.M, Kovach, A, Li, J, Yong, E.-L, Xu, H.E. | | Deposit date: | 2010-06-22 | | Release date: | 2010-08-25 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Identification and mechanism of ABA receptor antagonism.
Nat.Struct.Mol.Biol., 17, 2010
|
|
5TXP
 
 | | STRUCTURE OF Q151M complex (A62V, V75I, F77L, F116Y, Q151M) mutant HIV-1 REVERSE TRANSCRIPTASE (RT) TERNARY COMPLEX WITH A DOUBLE STRANDED DNA AND AN INCOMING DDATP | | Descriptor: | 1,2-ETHANEDIOL, 2',3'-dideoxyadenosine triphosphate, DNA (5'-D(*CP*AP*GP*TP*CP*CP*CP*TP*GP*TP*TP*CP*GP*GP*(MRG)P*CP*GP*CP*CP*G)-3'), ... | | Authors: | Das, K, Martinez, S.M, Arnold, E. | | Deposit date: | 2016-11-17 | | Release date: | 2017-04-05 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural Insights into HIV Reverse Transcriptase Mutations Q151M and Q151M Complex That Confer Multinucleoside Drug Resistance.
Antimicrob. Agents Chemother., 61, 2017
|
|
4Y8H
 
 | | Yeast 20S proteasome in complex with N3-APAL-ep | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Huber, E.M, Groll, M. | | Deposit date: | 2015-02-16 | | Release date: | 2015-06-17 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Systematic Analyses of Substrate Preferences of 20S Proteasomes Using Peptidic Epoxyketone Inhibitors.
J.Am.Chem.Soc., 137, 2015
|
|
6QRR
 
 | | X-ray radiation dose series on xylose isomerase - 0.13 MGy | | Descriptor: | 1,2-ETHANEDIOL, ISOPROPYL ALCOHOL, MAGNESIUM ION, ... | | Authors: | Taberman, H, Bury, C.S, van der Woerd, M.J, Snell, E.H, Garman, E.F. | | Deposit date: | 2019-02-19 | | Release date: | 2019-07-17 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.096 Å) | | Cite: | Structural knowledge or X-ray damage? A case study on xylose isomerase illustrating both.
J.Synchrotron Radiat., 26, 2019
|
|
6QRV
 
 | | X-ray radiation dose series on xylose isomerase - 2.63 MGy | | Descriptor: | 1,2-ETHANEDIOL, ISOPROPYL ALCOHOL, MAGNESIUM ION, ... | | Authors: | Taberman, H, Bury, C.S, van der Woerd, M.J, Snell, E.H, Garman, E.F. | | Deposit date: | 2019-02-19 | | Release date: | 2019-07-17 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.17 Å) | | Cite: | Structural knowledge or X-ray damage? A case study on xylose isomerase illustrating both.
J.Synchrotron Radiat., 26, 2019
|
|
6QSU
 
 | | Helicobacter pylori urease with BME bound in the active site | | Descriptor: | BETA-MERCAPTOETHANOL, NICKEL (II) ION, Urease subunit alpha, ... | | Authors: | Luecke, H, Cunha, E. | | Deposit date: | 2019-02-22 | | Release date: | 2021-01-20 | | Last modified: | 2021-02-03 | | Method: | ELECTRON MICROSCOPY (2.4 Å) | | Cite: | Cryo-EM structure of Helicobacter pylori urease with an inhibitor in the active site at 2.0 angstrom resolution.
Nat Commun, 12, 2021
|
|
3NCB
 
 | | A mutant human Prolactin receptor antagonist H180A in complex with the extracellular domain of the human prolactin receptor | | Descriptor: | CARBONATE ION, CHLORIDE ION, Prolactin, ... | | Authors: | Kulkarni, M.V, Tettamanzi, M.C, Murphy, J.W, Keeler, C, Myszka, D.G, Chayen, N.E, Lolis, E.J, Hodsdon, M.E. | | Deposit date: | 2010-06-04 | | Release date: | 2010-09-29 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Two Independent Histidines, One in Human Prolactin and One in Its Receptor, Are Critical for pH-dependent Receptor Recognition and Activation.
J.Biol.Chem., 285, 2010
|
|
1C9T
 
 | | COMPLEX OF BDELLASTASIN WITH BOVINE TRYPSIN | | Descriptor: | BDELLASTASIN, TRYPSIN | | Authors: | Rester, U, Bode, W, Moser, M, Parry, M.A, Huber, R, Auerswald, E. | | Deposit date: | 1999-08-03 | | Release date: | 2000-08-03 | | Last modified: | 2017-10-04 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structure of the complex of the antistasin-type inhibitor bdellastasin with trypsin and modelling of the bdellastasin-microplasmin system.
J.Mol.Biol., 293, 1999
|
|
8QU9
 
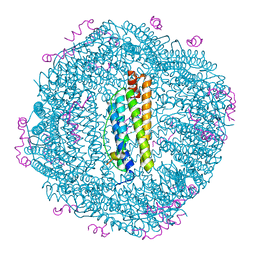 | | Structure of the NCOA4 (Nuclear Receptor Coactivator 4)-FTH1 (H-Ferritin) complex | | Descriptor: | FE (III) ION, Ferritin heavy chain, NCOA4 (Nuclear Receptor Coactivator 4) | | Authors: | Hoelzgen, F, Klukin, E, Zalk, R, Shahar, A, Cohen-Schwartz, S, Frank, G.A. | | Deposit date: | 2023-10-15 | | Release date: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (2.88 Å) | | Cite: | Structural basis for the intracellular regulation of ferritin degradation.
Nat Commun, 15, 2024
|
|
5MV1
 
 | | Crystal structure of the E protein of the Japanese encephalitis virulent virus | | Descriptor: | E protein | | Authors: | Liu, X, Zhao, X, Na, R, Li, L, Warkentin, E, Witt, J, Lu, X, Wei, Y, Peng, G, Li, Y, Wang, J. | | Deposit date: | 2017-01-14 | | Release date: | 2018-05-23 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | The structure differences of Japanese encephalitis virus SA14 and SA14-14-2 E proteins elucidate the virulence attenuation mechanism.
Protein Cell, 10, 2019
|
|
6Z1S
 
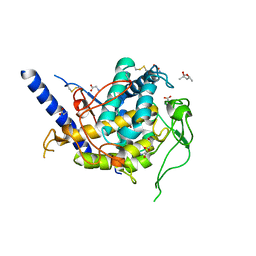 | | Structure of Polyphenol Oxidase (mutant G292N) from Thermothelomyces thermophila | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Dimarogona, M, Nikolaivits, E, Valmas, A, Topakas, E. | | Deposit date: | 2020-05-14 | | Release date: | 2021-03-24 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Considerations Regarding Activity Determinants of Fungal Polyphenol Oxidases Based on Mutational and Structural Studies.
Appl.Environ.Microbiol., 87, 2021
|
|
4ZHX
 
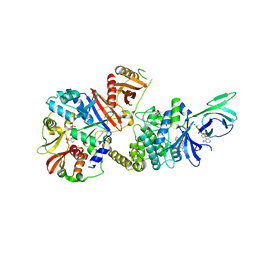 | | Novel binding site for allosteric activation of AMPK | | Descriptor: | (5S,6R,7R,9R,13cR,14R,16aS)-6-methoxy-5-methyl-7-(methylamino)-6,7,8,9,14,15,16,16a-octahydro-5H,13cH-5,9-epoxy-4b,9a,1 5-triazadibenzo[b,h]cyclonona[1,2,3,4-jkl]cyclopenta[e]-as-indacen-14-ol, 3-[4-(2-hydroxyphenyl)phenyl]-4-oxidanyl-6-oxidanylidene-7H-thieno[2,3-b]pyridine-5-carbonitrile, 5'-AMP-activated protein kinase catalytic subunit alpha-2, ... | | Authors: | Langendorf, C.G, Ngoei, K.R, Issa, S.M.A, Ling, N, Gorman, M.A, Parker, M.W, Sakamoto, K, Scott, J.W, Oakhill, J.S, Kemp, B.E. | | Deposit date: | 2015-04-27 | | Release date: | 2016-03-09 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.99 Å) | | Cite: | Structural basis of allosteric and synergistic activation of AMPK by furan-2-phosphonic derivative C2 binding.
Nat Commun, 7, 2016
|
|
6S52
 
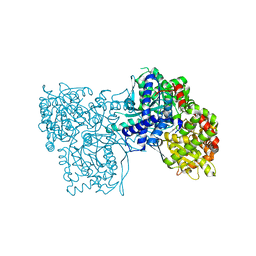 | | The crystal structure of glycogen phosphorylase in complex with 14 | | Descriptor: | (2~{R},3~{S},4~{S},5~{R},6~{R})-2-(hydroxymethyl)-6-(5-phenyl-1,2,3,4-tetrazol-2-yl)oxane-3,4,5-triol, Glycogen phosphorylase, muscle form, ... | | Authors: | Kyriakis, E, Solovou, T.G.A, Papaioannou, O.S.E, Skamnaki, V.T, Leonidas, D.D. | | Deposit date: | 2019-06-29 | | Release date: | 2020-02-19 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | The architecture of hydrogen and sulfur sigma-hole interactions explain differences in the inhibitory potency of C-beta-d-glucopyranosyl thiazoles, imidazoles and an N-beta-d glucopyranosyl tetrazole for human liver glycogen phosphorylase and offer new insights to structure-based design.
Bioorg.Med.Chem., 28, 2020
|
|
3NCF
 
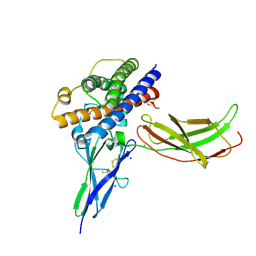 | | A mutant human Prolactin receptor antagonist H30A in complex with the mutant extracellular domain H188A of the human prolactin receptor | | Descriptor: | CHLORIDE ION, Prolactin, Prolactin receptor, ... | | Authors: | Kulkarni, M.V, Tettamanzi, M.C, Murphy, J.W, Keeler, C, Myszka, D.G, Chayen, N.E, Lolis, E.J, Hodsdon, M.E. | | Deposit date: | 2010-06-04 | | Release date: | 2010-09-29 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Two Independent Histidines, One in Human Prolactin and One in Its Receptor, Are Critical for pH-dependent Receptor Recognition and Activation.
J.Biol.Chem., 285, 2010
|
|
4ZKH
 
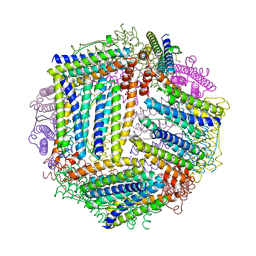 | |
6NPI
 
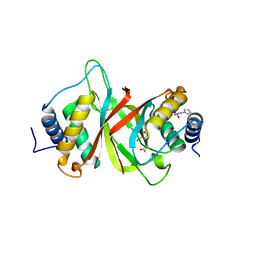 | | Crystal structure of Epstein-Barr Virus Nuclear Antigen-1, EBNA1, bound to fragments | | Descriptor: | ({2-[(4-bromo-5-methyl-1,2-oxazol-3-yl)amino]-2-oxoethyl}sulfanyl)acetic acid, 2-pyrrol-1-ylbenzoic acid, Epstein-Barr nuclear antigen 1 | | Authors: | Messick, T.E. | | Deposit date: | 2019-01-17 | | Release date: | 2019-03-20 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.501 Å) | | Cite: | Structure-based design of small-molecule inhibitors of EBNA1 DNA binding blocks Epstein-Barr virus latent infection and tumor growth.
Sci Transl Med, 11, 2019
|
|
6WRO
 
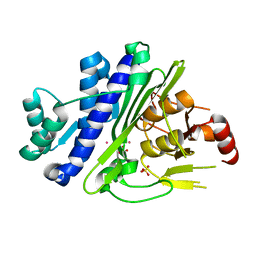 | |
4U9P
 
 | | Structure of the methanofuran/methanopterin biosynthetic enzyme MJ1099 from Methanocaldococcus jannaschii | | Descriptor: | GLYCEROL, UPF0264 protein MJ1099 | | Authors: | Bobik, T.A, Morales, E, Shin, A, Cascio, D, Sawaya, M.R, Arbing, M, Rasche, M.E. | | Deposit date: | 2014-08-06 | | Release date: | 2014-11-12 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure of the methanofuran/methanopterin-biosynthetic enzyme MJ1099 from Methanocaldococcus jannaschii.
Acta Crystallogr.,Sect.F, 70, 2014
|
|
6ZJA
 
 | | Helicobacter pylori urease with inhibitor bound in the active site | | Descriptor: | 2-{[1-(3,5-dimethylphenyl)-1H-imidazol-2-yl]sulfanyl}-N-hydroxyacetamide, NICKEL (II) ION, Urease subunit alpha, ... | | Authors: | Luecke, H, Cunha, E. | | Deposit date: | 2020-06-28 | | Release date: | 2020-12-23 | | Last modified: | 2021-02-03 | | Method: | ELECTRON MICROSCOPY (2 Å) | | Cite: | Cryo-EM structure of Helicobacter pylori urease with an inhibitor in the active site at 2.0 angstrom resolution.
Nat Commun, 12, 2021
|
|
6OFF
 
 | |
1AGM
 
 | | Refined structure for the complex of acarbose with glucoamylase from Aspergillus awamori var. x100 to 2.4 angstroms resolution | | Descriptor: | 4,6-dideoxy-4-{[(1S,4R,5S,6S)-4,5,6-trihydroxy-3-(hydroxymethyl)cyclohex-2-en-1-yl]amino}-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, GLUCOAMYLASE-471, alpha-D-mannopyranose, ... | | Authors: | Aleshin, A.E, Firsov, L.M, Honzatko, R.B. | | Deposit date: | 1994-05-13 | | Release date: | 1994-09-30 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Refined structure for the complex of acarbose with glucoamylase from Aspergillus awamori var. X100 to 2.4-A resolution.
J.Biol.Chem., 269, 1994
|
|
