1X9F
 
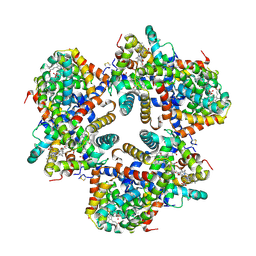 | | Hemoglobin Dodecamer from Lumbricus Erythrocruorin | | Descriptor: | CARBON MONOXIDE, Globin II, extracellular, ... | | Authors: | Strand, K, Knapp, J.E, Bhyravbhatla, B, Royer Jr, W.E. | | Deposit date: | 2004-08-20 | | Release date: | 2004-11-30 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of the hemoglobin dodecamer from lumbricus erythrocruorin: allosteric core of giant annelid respiratory complexes
J.Mol.Biol., 344, 2004
|
|
8CO3
 
 | |
6UWO
 
 | |
8CIB
 
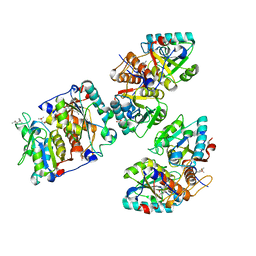 | | Structural and functional analysis of the Pseudomonas aeruginosa PA1677 protein | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, ACETATE ION, Cysteine hydrolase, ... | | Authors: | Sonnleitner, E, Brear, P, Luisi, B.F, Blasi, U. | | Deposit date: | 2023-02-09 | | Release date: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Catabolite repression control protein antagonist, a novel player in Pseudomonas aeruginosa carbon catabolite repression control.
Front Microbiol, 14, 2023
|
|
5FNU
 
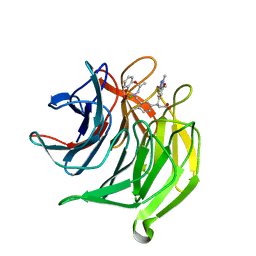 | | Structure of the Keap1 Kelch domain in complex with a small molecule inhibitor. | | Descriptor: | (3S)-3-(7-methoxy-1-methyl-1H-benzo[d][1,2,3]triazol-5-yl)-3-(4-methyl-3-(((R)-4-methyl-1,1-dioxido-3,4-dihydro-2H-benzo[b][1,4,5]oxathiazepin-2-yl)methyl)phenyl)propanoic acid, CHLORIDE ION, KELCH-LIKE ECH-ASSOCIATED PROTEIN 1 | | Authors: | Davies, T.G, Wixted, W.E, Coyle, J.E, Griffiths-Jones, C, Hearn, K, McMenamin, R, Norton, D, Rich, S.J, Richardson, C, Saxty, G, Willems, H.M.G, Woolford, A.J, Cottom, J.E, Kou, J, Yonchuk, J.G, Feldser, H.G, Sanchez, Y, Foley, J.P, Bolognese, B.J, Logan, G, Podolin, P.L, Yan, H, Callahan, J.F, Heightman, T.D, Kerns, J.K. | | Deposit date: | 2015-11-16 | | Release date: | 2016-04-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Mono-Acidic Inhibitors of the Kelch-Like Ech-Associated Protein 1 : Nuclear Factor Erythroid 2-Related Factor 2 (Keap1:Nrf2) Protein-Protein Interaction with High Cell Potency Identified by Fragment-Based Discovery.
J.Med.Chem., 59, 2016
|
|
8CBN
 
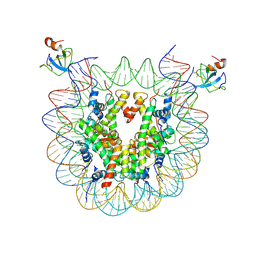 | | structure of LEDGF/p75 PWWP domain bound to the H3K36 trimethylated dinucleosome | | Descriptor: | Histone H2A, Histone H2B 1.1, Histone H3, ... | | Authors: | Koutna, E, Kouba, T, Novacek, J, Veverka, V. | | Deposit date: | 2023-01-25 | | Release date: | 2023-12-27 | | Method: | ELECTRON MICROSCOPY (3.34 Å) | | Cite: | Multivalency of nucleosome recognition by LEDGF.
Nucleic Acids Res., 51, 2023
|
|
6Y5C
 
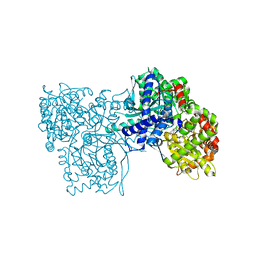 | | The crystal structure of glycogen phosphorylase in complex with 52 | | Descriptor: | 2-(4-methylphenyl)-5,7-bis(oxidanyl)chromen-4-one, Glycogen phosphorylase, muscle form | | Authors: | Kyriakis, E, Koulas, S.M, Skamnaki, V.T, Leonidas, D.D. | | Deposit date: | 2020-02-25 | | Release date: | 2020-08-19 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Synthetic flavonoid derivatives targeting the glycogen phosphorylase inhibitor site: QM/MM-PBSA motivated synthesis of substituted 5,7-dihydroxyflavones, crystallography, in vitro kinetics and ex-vivo cellular experiments reveal novel potent inhibitors.
Bioorg.Chem., 102, 2020
|
|
4XYE
 
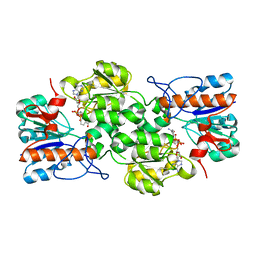 | | GRANULICELLA M. FORMATE DEHYDROGENASE (FDH) IN COMPLEX WITH NAD(+) | | Descriptor: | Formate dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Cendron, L, Fogal, S, Beneventi, E, Bergantino, E. | | Deposit date: | 2015-02-02 | | Release date: | 2015-07-08 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis for double cofactor specificity in a new formate dehydrogenase from the acidobacterium Granulicella mallensis MP5ACTX8.
Appl.Microbiol.Biotechnol., 99, 2015
|
|
8BZM
 
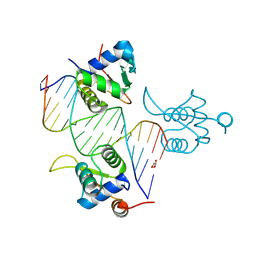 | | FOXK1-ELF1-heterodimer bound to DNA | | Descriptor: | DNA, ETS-related transcription factor Elf-1, Forkhead box protein K1, ... | | Authors: | Morgunova, E, Popov, A, Yin, Y, Taipale, J. | | Deposit date: | 2022-12-15 | | Release date: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | FOXK1-ELF1_heterodimer bound to DNA
To Be Published
|
|
3IO8
 
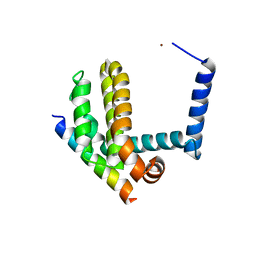 | | BimL12F in complex with Bcl-xL | | Descriptor: | Bcl-2-like protein 1, Bcl-2-like protein 11, ZINC ION | | Authors: | Colman, P.M, Lee, E.F, Fairlie, W.D, Smith, B.J, Czabotar, P.E, Yang, H, Sleebs, B.E, Lessene, G. | | Deposit date: | 2009-08-14 | | Release date: | 2009-09-01 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Conformational changes in Bcl-2 pro-survival proteins determine their capacity to bind ligands.
J.Biol.Chem., 284, 2009
|
|
3IOQ
 
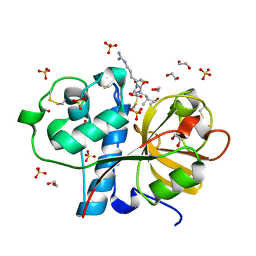 | | Crystal structure of the Carica candamarcensis cysteine protease CMS1MS2 in complex with E-64. | | Descriptor: | 1,2-ETHANEDIOL, CMS1MS2, N-[N-[1-HYDROXYCARBOXYETHYL-CARBONYL]LEUCYLAMINO-BUTYL]-GUANIDINE, ... | | Authors: | Gomes, M.T.R, Teixeira, R.D, Salas, C.E, Nagem, R.A.P. | | Deposit date: | 2009-08-14 | | Release date: | 2010-02-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Crystal structure of the Carica candamarcensis cysteine protease CMS1MS2 in complex with E-64
To be Published
|
|
6BT9
 
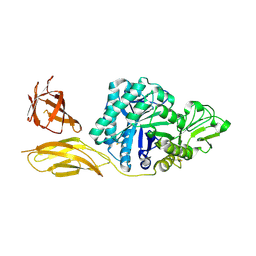 | | Chitinase ChiA74 from Bacillus thuringiensis | | Descriptor: | CALCIUM ION, Chitinase | | Authors: | Juarez-Hernandez, E, Brieba, L.G, Torres-Larios, A, Jimenez-Sandoval, P, Barboza-Corona, J. | | Deposit date: | 2017-12-05 | | Release date: | 2019-03-06 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | The crystal structure of the chitinase ChiA74 of Bacillus thuringiensis has a multidomain assembly.
Sci Rep, 9, 2019
|
|
6I5N
 
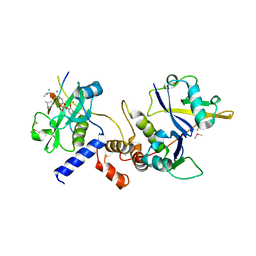 | | Crystal structure of SOCS2:Elongin C:Elongin B in complex with growth hormone receptor peptide | | Descriptor: | COBALT (II) ION, Elongin-B, Elongin-C, ... | | Authors: | Kung, W.W, Ramachandran, S, Makukhin, N, Bruno, E, Ciulli, A. | | Deposit date: | 2018-11-14 | | Release date: | 2019-05-29 | | Last modified: | 2019-06-19 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Structural insights into substrate recognition by the SOCS2 E3 ubiquitin ligase.
Nat Commun, 10, 2019
|
|
6I9E
 
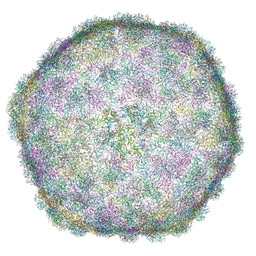 | | Thermophage P23-45 empty expanded capsid | | Descriptor: | Auxiliary protein, Major head protein | | Authors: | Bayfield, O.W, Klimuk, E, Winkler, D.C, Hesketh, E.L, Chechik, M, Cheng, N, Dykeman, E.C, Minakhin, L, Ranson, N.A, Severinov, K, Steven, A.C, Antson, A.A. | | Deposit date: | 2018-11-23 | | Release date: | 2019-02-06 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.74 Å) | | Cite: | Cryo-EM structure and in vitro DNA packaging of a thermophilic virus with supersized T=7 capsids.
Proc. Natl. Acad. Sci. U.S.A., 116, 2019
|
|
5FNT
 
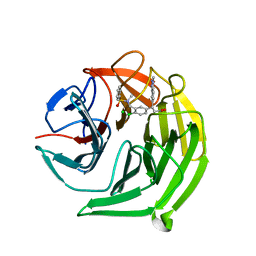 | | Structure of the Keap1 Kelch domain in complex with a small molecule inhibitor. | | Descriptor: | (3S)-3-{4-Chloro-3-[(N-methylbenzenesulfonamido) methyl]phenyl}-3-(1-methyl-1H-1,2,3-benzotriazol-5-yl)propanoic acid, CHLORIDE ION, KELCH-LIKE ECH-ASSOCIATED PROTEIN 1 | | Authors: | Davies, T.G, Wixted, W.E, Coyle, J.E, Griffiths-Jones, C, Hearn, K, McMenamin, R, Norton, D, Rich, S.J, Richardson, C, Saxty, G, Willems, H.M.G, Woolford, A.J, Cottom, J.E, Kou, J, Yonchuk, J.G, Feldser, H.G, Sanchez, Y, Foley, J.P, Bolognese, B.J, Logan, G, Podolin, P.L, Yan, H, Callahan, J.F, Heightman, T.D, Kerns, J.K. | | Deposit date: | 2015-11-16 | | Release date: | 2016-04-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Mono-Acidic Inhibitors of the Kelch-Like Ech-Associated Protein 1 : Nuclear Factor Erythroid 2-Related Factor 2 (Keap1:Nrf2) Protein-Protein Interaction with High Cell Potency Identified by Fragment-Based Discovery.
J.Med.Chem., 59, 2016
|
|
5FZJ
 
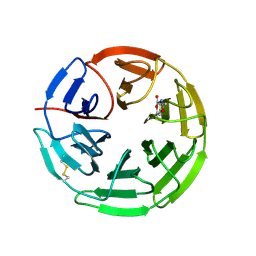 | | Structure of the Keap1 Kelch domain in complex with a small molecule inhibitor. | | Descriptor: | 2,6-DIMETHYL-4H-PYRANO[3,4-D][1,3]OXAZOL-4-ONE, KELCH-LIKE ECH-ASSOCIATED PROTEIN 1 | | Authors: | Davies, T.G, Wixted, W.E, Coyle, J.E, Griffiths-Jones, C, Hearn, K, McMenamin, R, Norton, D, Rich, S.J, Richardson, C, Saxty, G, Willems, H.M.G, Woolford, A.J, Cottom, J.E, Kou, J, Yonchuk, J.G, Feldser, H.G, Sanchez, Y, Foley, J.P, Bolognese, B.J, Logan, G, Podolin, P.L, Yan, H, Callahan, J.F, Heightman, T.D, Kerns, J.K. | | Deposit date: | 2016-03-14 | | Release date: | 2016-04-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Mono-Acidic Inhibitors of the Kelch-Like Ech-Associated Protein 1 : Nuclear Factor Erythroid 2-Related Factor 2 (Keap1:Nrf2) Protein-Protein Interaction with High Cell Potency Identified by Fragment-Based Discovery.
J.Med.Chem., 59, 2016
|
|
8P1H
 
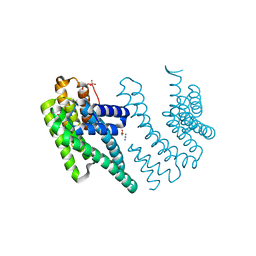 | | Crystal structure of the chimera of human 14-3-3 zeta and phosphorylated cytoplasmic loop fragment of the alpha7 acetylcholine receptor | | Descriptor: | 1,2-ETHANEDIOL, AZIDE ION, BENZOIC ACID, ... | | Authors: | Boyko, K.M, Kapitonova, A.A, Tugaeva, K.V, Varfolomeeva, L.A, Lyukmanova, E.N, Sluchanko, N.N. | | Deposit date: | 2023-05-12 | | Release date: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structure reveals canonical recognition of the phosphorylated cytoplasmic loop of human alpha7 nicotinic acetylcholine receptor by 14-3-3 protein.
Biochem.Biophys.Res.Commun., 682, 2023
|
|
6FFL
 
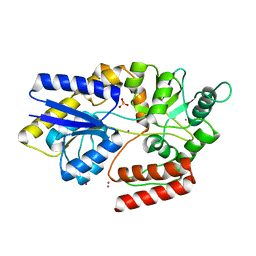 | | Maltose/maltodextrin-binding domain MalE from Bdellovibrio bacteriovorus bound to maltotriose | | Descriptor: | Maltose/maltodextrin transport permease homologue, PLATINUM (II) ION, SULFATE ION, ... | | Authors: | Licht, A, Werther, T, Bommer, M, Neumann, K, Schneider, E. | | Deposit date: | 2018-01-08 | | Release date: | 2018-01-24 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.707 Å) | | Cite: | Structural and functional characterization of a maltose/maltodextrin ABC transporter comprising a single solute binding domain (MalE) fused to the transmembrane subunit MalF.
Res. Microbiol., 170, 2018
|
|
6SBQ
 
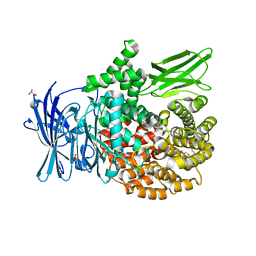 | | The crystal structure of PfA-M1 in complex with 7-amino-4-phenyl-5,7,8,9-tetrahydrobenzocyclohepten-6-one | | Descriptor: | DI(HYDROXYETHYL)ETHER, M1-family alanyl aminopeptidase, MALONATE ION, ... | | Authors: | Salomon, E, Schmitt, M, Mouray, E, McEwen, A.G, Torchy, M, Poussin-Courmontagne, P, Alavi, S, Tarnus, C, Cavarelli, J, Florent, I, Albrecht, S. | | Deposit date: | 2019-07-22 | | Release date: | 2020-03-25 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.33 Å) | | Cite: | Aminobenzosuberone derivatives as PfA-M1 inhibitors: Molecular recognition and antiplasmodial evaluation.
Bioorg.Chem., 98, 2020
|
|
6SDU
 
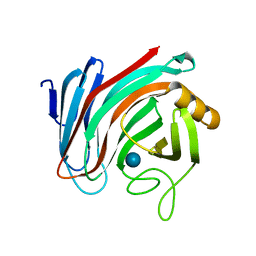 | | Xyloglucanase domain of NopAA, a type three effector from Sinorhizobium fredii in complex with cellobiose | | Descriptor: | Type III effector NopAA, beta-D-glucopyranose, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Dorival, D, Philys, S, Guintini, E, Brailly, R, de Ruyck, J, Czjzek, M, Biondi, E. | | Deposit date: | 2019-07-29 | | Release date: | 2020-07-08 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural and enzymatic characterisation of the Type III effector NopAA (=GunA) from Sinorhizobium fredii USDA257 reveals a Xyloglucan hydrolase activity.
Sci Rep, 10, 2020
|
|
8OYX
 
 | | De novo designed soluble GPCR-like fold GLF_18 | | Descriptor: | De novo designed soluble GPCR-like protein, PHOSPHATE ION | | Authors: | Pacesa, M, Correia, B.E. | | Deposit date: | 2023-05-05 | | Release date: | 2023-10-18 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Computational design of soluble functional analogues of integral membrane proteins.
Biorxiv, 2024
|
|
7B8A
 
 | | Notum-Fragment 110 | | Descriptor: | 1,2-ETHANEDIOL, 1-cyclohexyl-3-(2-pyridin-4-ylethyl)urea, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhao, Y, Jonees, E.Y. | | Deposit date: | 2020-12-12 | | Release date: | 2022-01-12 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.23 Å) | | Cite: | Notum Fragment Screen
To Be Published
|
|
6FHN
 
 | | Structural dynamics and catalytic properties of a multi-modular xanthanase (Pt derivative) | | Descriptor: | 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Moroz, O.V, Jensen, P.F, McDonald, S.P, McGregor, N, Blagova, E, Comamala, G, Segura, D.R, Anderson, L, Vasu, S.M, Rao, V.P, Giger, L, Monrad, R.N, Svendsen, A, Nielsen, J.E, Henrissat, B, Davies, G.J, Brumer, H, Rand, K, Wilson, K.S. | | Deposit date: | 2018-01-15 | | Release date: | 2018-08-29 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Dynamics and Catalytic Properties of a Multimodular Xanthanase
Acs Catalysis, 2018
|
|
5DKQ
 
 | | Crystal Structure of Calcium-loaded S100B bound to SBi4214 | | Descriptor: | 2,2'-[pentane-1,5-diylbis(oxybenzene-4,1-diyl)]di-1,4,5,6-tetrahydropyrimidine, CALCIUM ION, Protein S100-B | | Authors: | Cavalier, M.C, Ansari, M.I, Pierce, A.D, Wilder, P.T, McKnight, L.E, Raman, E.P, Neau, D.B, Bezawada, P, Alasady, M.J, Varney, K.M, Toth, E.A, MacKerell Jr, A.D, Coop, A, Weber, D.J. | | Deposit date: | 2015-09-03 | | Release date: | 2016-01-20 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.591 Å) | | Cite: | Small Molecule Inhibitors of Ca(2+)-S100B Reveal Two Protein Conformations.
J.Med.Chem., 59, 2016
|
|
4Y6N
 
 | | Crystal structure of glucosyl-3-phosphoglycerate synthase from Mycobacterium tuberculosis in complex with Mn2+, uridine-diphosphate-glucose (UDP-Glc) and phosphoglyceric acid (PGA) - GpgS Mn2+ UDP-Glc PGA-1 | | Descriptor: | 1,2-ETHANEDIOL, 3-PHOSPHOGLYCERIC ACID, Glucosyl-3-phosphoglycerate synthase, ... | | Authors: | Albesa-Jove, D, Rodrigo-Unzueta, A, Cifuente, J.O, Urresti, S, Comino, N, Sancho-Vaello, E, Guerin, M.E. | | Deposit date: | 2015-02-13 | | Release date: | 2015-07-15 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.348 Å) | | Cite: | A Native Ternary Complex Trapped in a Crystal Reveals the Catalytic Mechanism of a Retaining Glycosyltransferase.
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
