5AQJ
 
 | | Fragment-based screening of HSP70 sheds light on the functional role of ATP-binding site residues | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 9H-purine-6,8-diamine, BAG FAMILY MOLECULAR CHAPERONE REGULATOR 1, ... | | Authors: | Jones, A.M, Westwood, I.M, Osborne, J.D, Matthews, T.P, Cheeseman, M.D, Rowlands, M.G, Jeganathan, F, Burke, R, Lee, D, Kadi, N, Liu, M, Richards, M, McAndrew, C, Yahya, N, Dobson, S.E, Jones, K, Workman, P, Collins, I, van Montfort, R.L.M. | | Deposit date: | 2015-09-22 | | Release date: | 2016-10-05 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | A fragment-based approach applied to a highly flexible target: Insights and challenges towards the inhibition of HSP70 isoforms.
Sci Rep, 6, 2016
|
|
7U0P
 
 | | SARS-Cov2 S protein structure in complex with neutralizing monoclonal antibody 002-S21F2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Patel, A, Ortlund, E. | | Deposit date: | 2022-02-18 | | Release date: | 2022-08-10 | | Last modified: | 2023-02-22 | | Method: | ELECTRON MICROSCOPY (3.76 Å) | | Cite: | Structural insights for neutralization of Omicron variants BA.1, BA.2, BA.4, and BA.5 by a broadly neutralizing SARS-CoV-2 antibody.
Sci Adv, 8, 2022
|
|
6X4W
 
 | | Mfd-bound E.coli RNA polymerase elongation complex - III state | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, DNA (64-MER), DNA-directed RNA polymerase subunit alpha, ... | | Authors: | Llewellyn, E, Chen, J, Kang, J.Y, Darst, S.A. | | Deposit date: | 2020-05-24 | | Release date: | 2021-02-03 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structural basis for transcription complex disruption by the Mfd translocase.
Elife, 10, 2021
|
|
8B1V
 
 | | Dihydroprecondylocarpine acetate synthase 2 from Tabernanthe iboga | | Descriptor: | Dihydroprecondylocarpine acetate synthase 2, ZINC ION, precondylocarpine acetate | | Authors: | Langley, C, Basquin, J, Caputi, L, O'Connor, S.E. | | Deposit date: | 2022-09-12 | | Release date: | 2022-10-19 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.882 Å) | | Cite: | Expansion of the Catalytic Repertoire of Alcohol Dehydrogenases in Plant Metabolism.
Angew.Chem.Int.Ed.Engl., 61, 2022
|
|
2L62
 
 | | Protein and metal cluster structure of the wheat metallothionein domain g-Ec-1. The second part of the puzzle. | | Descriptor: | EC protein I/II, ZINC ION | | Authors: | Loebus, J, Peroza, E.A, Bluethgen, N, Fox, T, Meyer-Klaucke, W, Zerbe, O, Freisinger, E. | | Deposit date: | 2010-11-12 | | Release date: | 2011-05-25 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Protein and metal cluster structure of the wheat metallothionein domain gamma-E(c)-1: the second part of the puzzle.
J.Biol.Inorg.Chem., 16, 2011
|
|
6S4K
 
 | | The crystal structure of glycogen phosphorylase in complex with 12 | | Descriptor: | (2~{R},3~{S},4~{S},5~{R},6~{R})-2-(hydroxymethyl)-6-(4-phenyl-1,3-thiazol-2-yl)oxane-3,4,5-triol, Glycogen phosphorylase, muscle form, ... | | Authors: | Kyriakis, E, Solovou, T.G.A, Papaioannou, O.S.E, Skamnaki, V.T, Leonidas, D.D. | | Deposit date: | 2019-06-28 | | Release date: | 2020-02-19 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | The architecture of hydrogen and sulfur sigma-hole interactions explain differences in the inhibitory potency of C-beta-d-glucopyranosyl thiazoles, imidazoles and an N-beta-d glucopyranosyl tetrazole for human liver glycogen phosphorylase and offer new insights to structure-based design.
Bioorg.Med.Chem., 28, 2020
|
|
8UT2
 
 | | Pre-fusion Measles virus fusion protein complexed with Fab 77 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Fusion glycoprotein F0, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zyla, D, Saphire, E.O. | | Deposit date: | 2023-10-30 | | Release date: | 2024-07-03 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (2.56 Å) | | Cite: | A neutralizing antibody prevents postfusion transition of measles virus fusion protein.
Science, 384, 2024
|
|
8UUP
 
 | | Structure of the Measles virus Fusion protein in the pre-fusion conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Fusion glycoprotein F0, ... | | Authors: | Zyla, D, Saphire, E.O. | | Deposit date: | 2023-11-01 | | Release date: | 2024-07-03 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (2.11 Å) | | Cite: | A neutralizing antibody prevents postfusion transition of measles virus fusion protein.
Science, 384, 2024
|
|
2NTZ
 
 | |
4YFX
 
 | | Escherichia coli RNA polymerase in complex with Myxopyronin B | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Molodtsov, V, Fleming, P.R, Eyermann, C.J, Ferguson, A.D, Foulk, M.A, McKinney, D.C, Masse, C.E, Buurman, E.T, Murakami, K.S. | | Deposit date: | 2015-02-25 | | Release date: | 2015-03-11 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.844 Å) | | Cite: | X-ray Crystal Structures of Escherichia coli RNA Polymerase with Switch Region Binding Inhibitors Enable Rational Design of Squaramides with an Improved Fraction Unbound to Human Plasma Protein.
J.Med.Chem., 58, 2015
|
|
5NC6
 
 | | Crystal structure of the polysaccharide deacetylase Bc1974 from Bacillus cereus in complex with (E)-N-hydroxy-3-(naphthalen-1-yl)prop-2-enamide | | Descriptor: | 1,2-ETHANEDIOL, 3-naphthalen-1-yl-~{N}-oxidanyl-propanamide, ACETATE ION, ... | | Authors: | Giastas, P, Andreou, A, Balomenou, S, Bouriotis, V, Eliopoulos, E.E. | | Deposit date: | 2017-03-03 | | Release date: | 2018-02-21 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structures of the Peptidoglycan N-Acetylglucosamine Deacetylase Bc1974 and Its Complexes with Zinc Metalloenzyme Inhibitors.
Biochemistry, 57, 2018
|
|
8UUQ
 
 | |
7U0Q
 
 | |
7U0X
 
 | |
4YXW
 
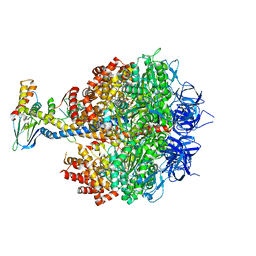 | | Bovine heart mitochondrial F1-ATPase inhibited by AMP-PNP and ADP in the presence of thiophosphate. | | Descriptor: | ATP synthase subunit alpha, mitochondrial, ATP synthase subunit beta, ... | | Authors: | Bason, J.V, Montgomery, M.G, Leslie, A.G.W, Walker, J.E. | | Deposit date: | 2015-03-23 | | Release date: | 2015-05-06 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | How release of phosphate from mammalian F1-ATPase generates a rotary substep.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
5KND
 
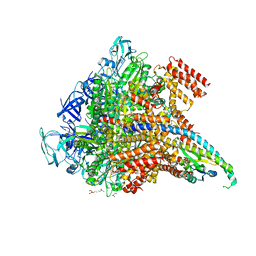 | | Crystal structure of the Pi-bound V1 complex | | Descriptor: | 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Suzuki, K, Mizutani, K, Maruyama, S, Shimono, K, Imai, F.L, Muneyuki, E, Kakinuma, Y, Ishizuka-Katsura, Y, Shirouzu, M, Yokoyama, S, Yamato, I, Murata, T. | | Deposit date: | 2016-06-28 | | Release date: | 2016-11-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.888 Å) | | Cite: | Crystal structures of the ATP-binding and ADP-release dwells of the V1 rotary motor
Nat Commun, 7, 2016
|
|
8ANQ
 
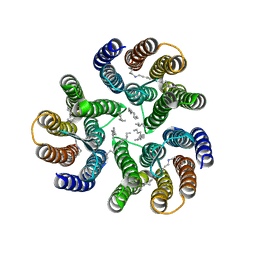 | | Crystal structure of the microbial rhodopsin from Sphingomonas paucimobilis (SpaR) | | Descriptor: | Bacteriorhodopsin, EICOSANE | | Authors: | Kovalev, K, Okhrimenko, I, Marin, E, Gordeliy, V. | | Deposit date: | 2022-08-05 | | Release date: | 2023-05-03 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Mirror proteorhodopsins.
Commun Chem, 6, 2023
|
|
5L8C
 
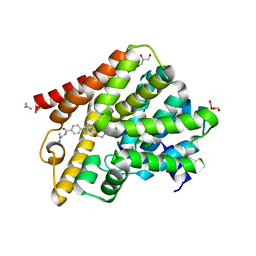 | | Crystal structure of T. brucei PDE-B1 catalytic domain with inhibitor NPD-039 | | Descriptor: | 4-[5-[(4~{a}~{R},8~{a}~{S})-4-oxidanylidene-3-propan-2-yl-4~{a},5,8,8~{a}-tetrahydrophthalazin-1-yl]-2-methoxy-phenyl]-~{N}-(2-azanyl-2-oxidanylidene-ethyl)benzamide, DI(HYDROXYETHYL)ETHER, FORMIC ACID, ... | | Authors: | Singh, A.K, Anthonyrajah, E.S, Brown, D.G. | | Deposit date: | 2016-06-07 | | Release date: | 2018-03-14 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Targeting a Subpocket in Trypanosoma brucei Phosphodiesterase B1 (TbrPDEB1) Enables the Structure-Based Discovery of Selective Inhibitors with Trypanocidal Activity.
J. Med. Chem., 61, 2018
|
|
1YPP
 
 | | ACID ANHYDRIDE HYDROLASE | | Descriptor: | INORGANIC PYROPHOSPHATASE, MANGANESE (II) ION, PHOSPHATE ION | | Authors: | Harutyunyan, E.H, Kuranova, I.P, Lamzin, V.S, Dauter, Z, Wilson, K.S. | | Deposit date: | 1996-05-29 | | Release date: | 1996-12-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | X-ray structure of yeast inorganic pyrophosphatase complexed with manganese and phosphate.
Eur.J.Biochem., 239, 1996
|
|
8SBD
 
 | | Cryo-EM structure of insulin amyloid-like fibril that is composed of two antiparallel protofilaments | | Descriptor: | Insulin A chain, Insulin B chain | | Authors: | Wang, L.W, Hall, C, Uchikawa, E, Chen, D.L, Choi, E, Zhang, X.W, Bai, X.C. | | Deposit date: | 2023-04-03 | | Release date: | 2023-08-30 | | Last modified: | 2023-09-27 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural basis of insulin fibrillation.
Sci Adv, 9, 2023
|
|
5AQR
 
 | | Fragment-based screening of HSP70 sheds light on the functional role of ATP-binding site residues | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 6-methoxyquinazolin-4-amine, BAG FAMILY MOLECULAR CHAPERONE REGULATOR 1, ... | | Authors: | Jones, A.M, Westwood, I.M, Osborne, J.D, Matthews, T.P, Cheeseman, M.D, Rowlands, M.G, Jeganathan, F, Burke, R, Lee, D, Kadi, N, Liu, M, Richards, M, McAndrew, C, Yahya, N, Dobson, S.E, Jones, K, Workman, P, Collins, I, van Montfort, R.L.M. | | Deposit date: | 2015-09-22 | | Release date: | 2016-10-05 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | A fragment-based approach applied to a highly flexible target: Insights and challenges towards the inhibition of HSP70 isoforms.
Sci Rep, 6, 2016
|
|
2NU6
 
 | | C123aA Mutant of E. coli Succinyl-CoA Synthetase | | Descriptor: | COENZYME A, SULFATE ION, Succinyl-CoA ligase [ADP-forming] subunit alpha, ... | | Authors: | Fraser, M.E. | | Deposit date: | 2006-11-08 | | Release date: | 2007-07-24 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Participation of Cys 123alpha of Escherichia coli Succinyl-CoA Synthetase in Catalysis
ACTA CRYSTALLOGR.,SECT.D, 63, 2007
|
|
5DXB
 
 | | Estrogen Receptor Alpha Ligand Binding Domain Y537S Mutant in Complex with Stapled Peptide SRC2-P1 and Estradiol | | Descriptor: | CHLORIDE ION, ESTRADIOL, Estrogen receptor, ... | | Authors: | Fanning, S.W, Speltz, T.E, Mayne, C.G, Tajkhorshid, E, Greene, G.L, Moore, T.W. | | Deposit date: | 2015-09-23 | | Release date: | 2016-07-27 | | Last modified: | 2020-02-19 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Stapled Peptides with gamma-Methylated Hydrocarbon Chains for the Estrogen Receptor/Coactivator Interaction.
Angew. Chem. Int. Ed. Engl., 55, 2016
|
|
2NU7
 
 | | C123aS Mutant of E. coli Succinyl-CoA Synthetase | | Descriptor: | COENZYME A, PHOSPHATE ION, SULFATE ION, ... | | Authors: | Fraser, M.E. | | Deposit date: | 2006-11-08 | | Release date: | 2007-07-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Participation of Cys 123alpha of Escherichia coli Succinyl-CoA Synthetase in Catalysis
ACTA CRYSTALLOGR.,SECT.D, 63, 2007
|
|
7GB8
 
 | | Group deposition SARS-CoV-2 main protease in complex with inhibitors from the COVID Moonshot -- Crystal Structure of SARS-CoV-2 main protease in complex with JAN-GHE-83b26c96-14 (Mpro-x10247) | | Descriptor: | 3C-like proteinase, DIMETHYL SULFOXIDE, N-(4-methylpyridin-3-yl)-2-[3-(trifluoromethyl)phenyl]acetamide | | Authors: | Fearon, D, Aimon, A, Aschenbrenner, J.C, Balcomb, B.H, Bertram, F.K.R, Brandao-Neto, J, Dias, A, Douangamath, A, Dunnett, L, Godoy, A.S, Gorrie-Stone, T.J, Koekemoer, L, Krojer, T, Lithgo, R.M, Lukacik, P, Marples, P.G, Mikolajek, H, Nelson, E, Owen, C.D, Powell, A.J, Rangel, V.L, Skyner, R, Strain-Damerell, C.M, Thompson, W, Tomlinson, C.W.E, Wild, C, Walsh, M.A, von Delft, F. | | Deposit date: | 2023-08-11 | | Release date: | 2023-11-08 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.957 Å) | | Cite: | Open science discovery of potent noncovalent SARS-CoV-2 main protease inhibitors.
Science, 382, 2023
|
|
