6B6X
 
 | | Crystal structure of Desulfovibrio vulgaris carbon monoxide dehydrogenase, dithionite-reduced (protein batch 2), canonical C-cluster | | Descriptor: | CHLORIDE ION, Carbon monoxide dehydrogenase, FE(4)-NI(1)-S(4) CLUSTER, ... | | Authors: | Wittenborn, E.C, Drennan, C.L. | | Deposit date: | 2017-10-03 | | Release date: | 2018-10-03 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Redox-dependent rearrangements of the NiFeS cluster of carbon monoxide dehydrogenase.
Elife, 7, 2018
|
|
4LF7
 
 | | Crystal Structure of 30S ribosomal subunit from Thermus thermophilus | | Descriptor: | 16S rRNA, MAGNESIUM ION, PAROMOMYCIN, ... | | Authors: | Demirci, H, Belardinelli, R, Carr, J, Murphy IV, F, Jogl, G, Dahlberg, A.E, Gregory, S.T. | | Deposit date: | 2013-06-26 | | Release date: | 2014-07-02 | | Method: | X-RAY DIFFRACTION (3.1484 Å) | | Cite: | Crystal Structure of 30S ribosomal subunit from Thermus thermophilus
To be Published, 2013
|
|
1B96
 
 | |
3MNR
 
 | | Crystal Structure of Benzamide SNX-1321 bound to Hsp90 | | Descriptor: | 2-[(3,4,5-trimethoxyphenyl)amino]-4-(2,6,6-trimethyl-4-oxo-4,5,6,7-tetrahydro-1H-indol-1-yl)benzamide, Heat shock protein HSP 90-alpha | | Authors: | Veal, J.M, Fadden, P, Huang, K.H, Rice, J, Hall, S.E, Haytstead, T.A. | | Deposit date: | 2010-04-22 | | Release date: | 2010-08-11 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Application of Chemoproteomics to Drug Discovery: Identification of a Clinical Candidate Targeting Hsp90.
Chem.Biol., 17, 2010
|
|
7KRA
 
 | | Cryo-EM structure of Saccharomyces cerevisiae ER membrane protein complex bound to Fab-DH4 in lipid nanodiscs | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ER membrane protein complex subunit 1, ER membrane protein complex subunit 2, ... | | Authors: | Miller-Vedam, L.E, Schirle Oakdale, N.S, Braeuning, B, Boydston, E.A, Sevillano, N, Popova, K.D, Bonnar, J.L, Shurtleff, M.J, Prabu, J.R, Stroud, R.M, Craik, C.S, Schulman, B.A, Weissman, J.S, Frost, A. | | Deposit date: | 2020-11-19 | | Release date: | 2020-12-02 | | Last modified: | 2020-12-09 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural and mechanistic basis of the EMC-dependent biogenesis of distinct transmembrane clients.
Elife, 9, 2020
|
|
1UM5
 
 | | Catalytic Antibody 21H3 with alcohol substrate | | Descriptor: | 1-PHENYLETHANOL, Antibody 21H3 H chain, Antibody 21H3 L chain | | Authors: | Beuscher IV, A.E, Reuter, J, Olson, A.J, Romesberg, F.E, Schultz, P.G, Wirsching, P, Janda, K.D, Lerner, R.A, Stevens, R.C. | | Deposit date: | 2003-09-23 | | Release date: | 2004-10-05 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural Studies of an Efficient Catalytic Antibody Operating by Ping-Pong and Induced Fit Mechanisms
To be Published
|
|
1FM4
 
 | | CRYSTAL STRUCTURE OF THE BIRCH POLLEN ALLERGEN BET V 1L | | Descriptor: | (3ALPHA,5BETA,12ALPHA)-3,12-DIHYDROXYCHOLAN-24-OIC ACID, MAJOR POLLEN ALLERGEN BET V 1-L | | Authors: | Markovic-Housley, Z, Degano, M, Lamba, D, von Roepenack-Lahaye, E, Clemens, S, Susani, M, Ferreira, F, Scheiner, O, Breiteneder, H. | | Deposit date: | 2000-08-16 | | Release date: | 2002-12-20 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Crystal Structure of a Hypoallergenic Isoform of the Major Birch Pollen Allergen Bet
v 1 and its Likely Biological Function as a Plant Steroid Carrier
J.Mol.Biol., 325, 2003
|
|
1ULI
 
 | | Biphenyl dioxygenase (BphA1A2) derived from Rhodococcus sp. strain RHA1 | | Descriptor: | FE (II) ION, FE2/S2 (INORGANIC) CLUSTER, biphenyl dioxygenase large subunit, ... | | Authors: | Furusawa, Y, Nagarajan, V, Masai, E, Tanokura, M, Fukuda, M, Senda, T. | | Deposit date: | 2003-09-12 | | Release date: | 2004-09-28 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structure of the Terminal Oxygenase Component of Biphenyl Dioxygenase Derived from Rhodococcus sp. Strain RHA1
J.Mol.Biol., 342, 2004
|
|
2JYQ
 
 | | NMR structure of the apo v-Src SH2 domain | | Descriptor: | Tyrosine-protein kinase transforming protein Src | | Authors: | Taylor, J.D, Ababou, A, Williams, M.A, Ladbury, J.E. | | Deposit date: | 2007-12-17 | | Release date: | 2008-06-24 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structure, dynamics, and binding thermodynamics of the v-Src SH2 domain: Implications for drug design
Proteins, 73, 2008
|
|
1UWF
 
 | | 1.7 A resolution structure of the receptor binding domain of the FimH adhesin from uropathogenic E. coli | | Descriptor: | FIMH PROTEIN, GLYCEROL, butyl alpha-D-mannopyranoside | | Authors: | Bouckaert, J, Berglund, J, Genst, E.D, Cools, L, Hung, C.-S, Wuhrer, M, Zavialov, A, Langermann, S, Hultgren, S, Wyns, L, Oscarson, S, Knight, S.D, De Greve, H. | | Deposit date: | 2004-02-05 | | Release date: | 2005-02-16 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Receptor Binding Studies Disclose a Novel Class of High-Affinity Inhibitors of the Escherichia Coli Fimh Adhesin.
Mol.Microbiol., 55, 2005
|
|
7KEY
 
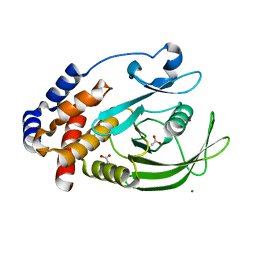 | | Protein Tyrosine Phosphatase 1B, Apo | | Descriptor: | ACETATE ION, MAGNESIUM ION, Tyrosine-protein phosphatase non-receptor type 1 | | Authors: | Battaile, K.P, Chirgadze, Y, Ruzanov, M, Romanov, V, Lam, K, Gordon, R, Lin, A, Lam, R, Pai, E, Chirgadze, N. | | Deposit date: | 2020-10-13 | | Release date: | 2022-01-19 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.771 Å) | | Cite: | Signal transfer in human protein tyrosine phosphatase PTP1B from allosteric inhibitor P00058.
J.Biomol.Struct.Dyn., 2021
|
|
1UXV
 
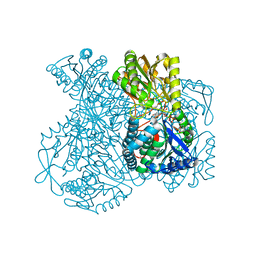 | | Structural basis for allosteric regulation and substrate specificity of the non-phosphorylating glyceraldehyde-3-phosphate dehydrogenase (GAPN) from Thermoproteus tenax | | Descriptor: | ADENOSINE MONOPHOSPHATE, GLYCERALDEHYDE-3-PHOSPHATE DEHYDROGENASE (NADP+), NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Lorentzen, E, Hensel, R, Pohl, E. | | Deposit date: | 2004-03-01 | | Release date: | 2004-08-05 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural Basis of Allosteric Regulation and Substrate Specificity of the Non-Phosphorylating Glyceraldehyde 3-Phosphate Dehydrogenase from Thermoproteus Tenax
J.Mol.Biol., 341, 2004
|
|
3RAF
 
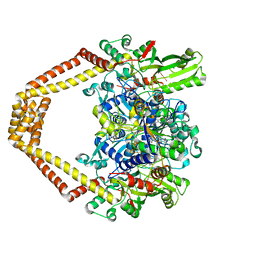 | | Quinazolinedione-DNA cleavage complex of type IV topoisomerase from S. pneumoniae | | Descriptor: | 3-amino-7-{(3R)-3-[(1S)-1-aminoethyl]pyrrolidin-1-yl}-1-cyclopropyl-6-fluoro-8-methylquinazoline-2,4(1H,3H)-dione, 5'-D(*CP*AP*TP*GP*AP*AP*T)-3', 5'-D(*CP*GP*TP*GP*CP*AP*T)-3', ... | | Authors: | Laponogov, I, Pan, X.-S, Veselkov, D.A, McAuley, K.E, Fisher, L.M, Sanderson, M.R. | | Deposit date: | 2011-03-28 | | Release date: | 2012-03-28 | | Method: | X-RAY DIFFRACTION (3.24 Å) | | Cite: | Inhibitor-stabilised cleavage complexes of topoisomerase IIa: structural analysis of drug-dependent inter- and intramolecular interactions
To be Published
|
|
1EFI
 
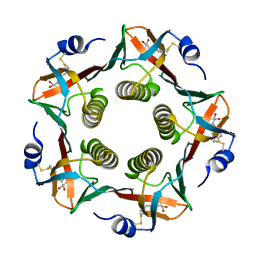 | |
5BRZ
 
 | | MAGE-A3 reactive TCR in complex with MAGE-A3 in HLA-A1 | | Descriptor: | Beta-2-microglobulin, GLU-VAL-ASP-PRO-ILE-GLY-HIS-LEU-TYR, HLA class I histocompatibility antigen, ... | | Authors: | Raman, M.C.C, Rizkallah, P.J, Simmons, R, Donnellan, Z, Dukes, J, Bossi, G, LeProvost, G, Mahon, T, Hickman, E, LomaX, M, Oates, J, Hassan, N, Vuidepot, A, Sami, M, Cole, D.K, Jakobsen, B.K. | | Deposit date: | 2015-06-01 | | Release date: | 2016-03-02 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.62 Å) | | Cite: | Direct molecular mimicry enables off-target cardiovascular toxicity by an enhanced affinity TCR designed for cancer immunotherapy.
Sci Rep, 6, 2016
|
|
9B9G
 
 | | Structure of the PI4KA complex bound to Calcineurin | | Descriptor: | CALCIUM ION, Calcineurin subunit B type 1, Hyccin, ... | | Authors: | Shaw, A.L, Suresh, S, Yip, C.K, Burke, J.E. | | Deposit date: | 2024-04-02 | | Release date: | 2024-09-11 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structure of calcineurin bound to PI4KA reveals dual interface in both PI4KA and FAM126A.
Structure, 2024
|
|
7L5C
 
 | | Structure of copper bound MEMO1 | | Descriptor: | 1,2-ETHANEDIOL, COPPER (I) ION, GLYCEROL, ... | | Authors: | Boniecki, M.T, Uhlemann, E.E, Dmitriev, O.Y. | | Deposit date: | 2020-12-21 | | Release date: | 2022-01-19 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | MEMO1 binds iron and modulates iron homeostasis in cancer cells.
Elife, 13, 2024
|
|
1FMD
 
 | | THE STRUCTURE AND ANTIGENICITY OF A TYPE C FOOT-AND-MOUTH DISEASE VIRUS | | Descriptor: | FOOT-AND-MOUTH DISEASE VIRUS (SUBUNIT VP1), FOOT-AND-MOUTH DISEASE VIRUS (SUBUNIT VP2), FOOT-AND-MOUTH DISEASE VIRUS (SUBUNIT VP3), ... | | Authors: | Lea, S, Fry, E, Stuart, D. | | Deposit date: | 1994-02-10 | | Release date: | 1994-08-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | The structure and antigenicity of a type C foot-and-mouth disease virus.
Structure, 2, 1994
|
|
4Y8O
 
 | |
3M1V
 
 | | Structural Insight into Methyl-Coenzyme M Reductase Chemistry using Coenzyme B Analogues | | Descriptor: | 1,2-ETHANEDIOL, 1-THIOETHANESULFONIC ACID, ACETATE ION, ... | | Authors: | Cedervall, P.E, Dey, M, Ragsdale, S.W, Wilmot, C.M. | | Deposit date: | 2010-03-05 | | Release date: | 2010-09-15 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structural insight into methyl-coenzyme M reductase chemistry using coenzyme B analogues.
Biochemistry, 49, 2010
|
|
1YTS
 
 | | A LIGAND-INDUCED CONFORMATIONAL CHANGE IN THE YERSINIA PROTEIN TYROSINE PHOSPHATASE | | Descriptor: | SULFATE ION, YERSINIA PROTEIN TYROSINE PHOSPHATASE | | Authors: | Schubert, H.L, Stuckey, J.A, Fauman, E.B, Dixon, J.E, Saper, M.A. | | Deposit date: | 1995-04-07 | | Release date: | 1995-07-10 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A ligand-induced conformational change in the Yersinia protein tyrosine phosphatase.
Protein Sci., 4, 1995
|
|
4N56
 
 | |
1NXF
 
 | |
4YEC
 
 | |
2JV6
 
 | | YF ED3 Protein NMR Structure | | Descriptor: | Envelope protein E | | Authors: | Volk, D.E, Gandham, S.H.A, May, F.J, Anderson, A, Barrett, A.D.T, Gorenstein, D.G. | | Deposit date: | 2007-09-12 | | Release date: | 2008-09-16 | | Last modified: | 2022-03-16 | | Method: | SOLUTION NMR | | Cite: | Yellow Fever Virus Envelope Protein Domain III: A Convergence of Structure and Phylogenetics
To be Published
|
|
