5KRK
 
 | | Crystal Structure of the ER-alpha Ligand-binding Domain (Y537S) in Complex with 4,4'-((5-bromo-2,3-dihydro-1H-inden-1-ylidene)methylene)diphenol | | Descriptor: | 4-[(5-bromanyl-2,3-dihydroinden-1-ylidene)-(4-hydroxyphenyl)methyl]phenol, Estrogen receptor, NCOA2 | | Authors: | Nwachukwu, J.C, Srinivasan, S, Bruno, N.E, Nowak, J, Kojetin, D.J, Elemento, O, Katzenellenbogen, J.A, Nettles, K.W. | | Deposit date: | 2016-07-07 | | Release date: | 2017-01-18 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.391 Å) | | Cite: | Systems Structural Biology Analysis of Ligand Effects on ER alpha Predicts Cellular Response to Environmental Estrogens and Anti-hormone Therapies.
Cell Chem Biol, 24, 2017
|
|
6YCY
 
 | | Plasmodium falciparum Myosin A full-length, post-rigor state | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Moussaoui, D, Robblee, J.P, Auguin, D, Krementsova, E.B, Robert-Paganin, J, Trybus, K.M, Houdusse, A. | | Deposit date: | 2020-03-19 | | Release date: | 2020-11-11 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Full-length Plasmodium falciparum myosin A and essential light chain PfELC structures provide new anti-malarial targets.
Elife, 9, 2020
|
|
7OQD
 
 | | A single sulfatase is required for metabolism of colonic mucin O-glycans and intestinal colonization by a symbiotic human gut bacterium (BT1636-S1_20) | | Descriptor: | 3-O-sulfo-beta-D-galactopyranose, Arylsulfatase, CALCIUM ION | | Authors: | Sofia de Jesus Vaz Luis, A, Basle, A, Martens, E.C, Cartmell, A. | | Deposit date: | 2021-06-03 | | Release date: | 2021-10-27 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A single sulfatase is required to access colonic mucin by a gut bacterium.
Nature, 598, 2021
|
|
6RZ8
 
 | | Crystal structure of the human cysteinyl leukotriene receptor 2 in complex with ONO-2080365 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (2~{S})-8-[[4-[4-[2,3-bis(fluoranyl)phenoxy]butoxy]-2-fluoranyl-phenyl]carbonylamino]-4-(4-oxidanyl-4-oxidanylidene-but yl)-2,3-dihydro-1,4-benzoxazine-2-carboxylic acid, Cysteinyl leukotriene receptor 2,Soluble cytochrome b562,Cysteinyl leukotriene receptor 2, ... | | Authors: | Gusach, A, Luginina, A, Marin, E, Brouillette, R.L, Besserer-Offroy, E, Longpre, J.M, Ishchenko, A, Popov, P, Fujimoto, T, Maruyama, T, Stauch, B, Ergasheva, M, Romanovskaya, D, Stepko, A, Kovalev, K, Shevtsov, M, Gordeliy, V, Han, G.W, Sarret, P, Katritch, V, Borshchevskiy, V, Mishin, A, Cherezov, V. | | Deposit date: | 2019-06-12 | | Release date: | 2019-12-11 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis of ligand selectivity and disease mutations in cysteinyl leukotriene receptors.
Nat Commun, 10, 2019
|
|
8P64
 
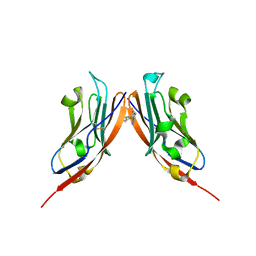 | | Co-crystal structure of PD-L1 with low molecular weight inhibitor | | Descriptor: | Programmed cell death 1 ligand 1, ~{N}-[[1-[(~{E})-2-(2-methyl-3-phenyl-phenyl)ethenyl]-1,2,3,4-tetrazol-5-yl]methyl]ethanamine | | Authors: | Plewka, J, Magiera-Mularz, K, van der Straat, R, Draijer, R, Surmiak, E, Butera, R, Land, L, Musielak, B, Domling, A. | | Deposit date: | 2023-05-25 | | Release date: | 2024-03-06 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (3.312 Å) | | Cite: | 1,5-Disubstituted tetrazoles as PD-1/PD-L1 antagonists.
Rsc Med Chem, 15, 2024
|
|
5EUQ
 
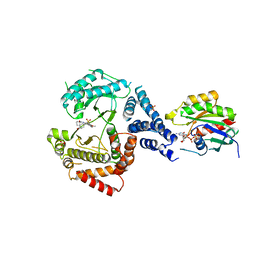 | |
8R1L
 
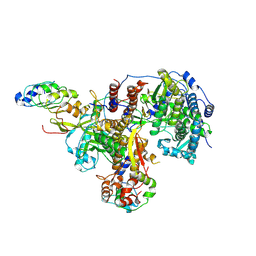 | | Structure of avian H5N1 influenza A polymerase in complex with human ANP32B. | | Descriptor: | Acidic leucine-rich nuclear phosphoprotein 32 family member B, Polymerase acidic protein, Polymerase basic protein 2, ... | | Authors: | Carrique, L, Staller, E, Keown, J.R, Fan, H, Fodor, E, Grimes, J.M. | | Deposit date: | 2023-11-02 | | Release date: | 2024-05-08 | | Last modified: | 2024-07-31 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structures of H5N1 influenza polymerase with ANP32B reveal mechanisms of genome replication and host adaptation.
Nat Commun, 15, 2024
|
|
4YSS
 
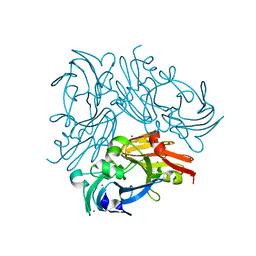 | | Structure of copper nitrite reductase from Geobacillus thermodenitrificans - 16.7 MGy | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, COPPER (II) ION, Nitrite reductase, ... | | Authors: | Fukuda, Y, Tse, K.M, Suzuki, M, Diedrichs, K, Hirata, K, Nakane, T, Sugahara, M, Nango, E, Tono, K, Joti, Y, Kameshima, T, Song, C, Hatsui, T, Yabashi, M, Nureki, O, Matsumura, H, Inoue, T, Iwata, S, Mizohata, E. | | Deposit date: | 2015-03-17 | | Release date: | 2016-02-24 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Redox-coupled structural changes in nitrite reductase revealed by serial femtosecond and microfocus crystallography
J.Biochem., 159, 2016
|
|
7P39
 
 | | 4,6-alpha-glucanotransferase GtfB from Limosilactobacillus reuteri NCC 2613 complexed with acarbose | | Descriptor: | 4,6-dideoxy-4-{[(1S,4R,5S,6S)-4,5,6-trihydroxy-3-(hydroxymethyl)cyclohex-2-en-1-yl]amino}-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-beta-D-glucopyranose, CALCIUM ION, Dextransucrase, ... | | Authors: | Pijning, T, te Poele, E, Gangoiti, J, Boerner, T, Dijkhuizen, L. | | Deposit date: | 2021-07-07 | | Release date: | 2021-11-03 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Insights into Broad-Specificity Starch Modification from the Crystal Structure of Limosilactobacillus Reuteri NCC 2613 4,6-alpha-Glucanotransferase GtfB.
J.Agric.Food Chem., 69, 2021
|
|
5DKR
 
 | | Crystal Structure of Calcium-loaded S100B bound to SBi29 | | Descriptor: | 2-[4-(4-carbamimidoylphenoxy)phenyl]-1H-indole-6-carboximidamide, CALCIUM ION, Protein S100-B | | Authors: | Cavalier, M.C, Ansari, M.I, Pierce, A.D, Wilder, P.T, McKnight, L.E, Raman, E.P, Neau, D.B, Bezawada, P, Alasady, M.J, Varney, K.M, Toth, E.A, MacKerell Jr, A.D, Coop, A, Weber, D.J. | | Deposit date: | 2015-09-03 | | Release date: | 2016-01-20 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.742 Å) | | Cite: | Small Molecule Inhibitors of Ca(2+)-S100B Reveal Two Protein Conformations.
J.Med.Chem., 59, 2016
|
|
7P38
 
 | | 4,6-alpha-glucanotransferase GtfB from Limosilactobacillus reuteri NCC 2613 | | Descriptor: | CALCIUM ION, Dextransucrase, GLYCEROL, ... | | Authors: | Pijning, T, te Poele, E, Gangoiti, J, Boerner, T, Dijkhuizen, L. | | Deposit date: | 2021-07-07 | | Release date: | 2021-11-03 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Insights into Broad-Specificity Starch Modification from the Crystal Structure of Limosilactobacillus Reuteri NCC 2613 4,6-alpha-Glucanotransferase GtfB.
J.Agric.Food Chem., 69, 2021
|
|
6GMB
 
 | | Structure of human hydroxyacid oxidase 1 bound with FMN and glycolate | | Descriptor: | 1,2-ETHANEDIOL, FLAVIN MONONUCLEOTIDE, GLYCOLIC ACID, ... | | Authors: | MacKinnon, S, Bezerra, G.A, Krojer, T, Smee, C, Arrowsmith, C.H, Edwards, E, Bountra, C, Oppermann, U, Brennan, P.E, Yue, W.W, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-05-24 | | Release date: | 2018-06-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Structure of human hydroxyacid oxidase 1 bound with FMN and glycolate
To Be Published
|
|
2WAB
 
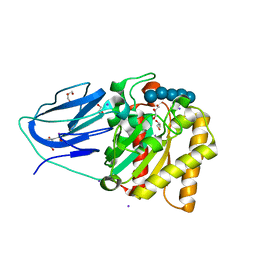 | | Structure of an active site mutant of a family two carbohydrate esterase from Clostridium thermocellum in complex with celluohexase | | Descriptor: | ENDOGLUCANASE E, GLYCEROL, IODIDE ION, ... | | Authors: | Montainer, C, Money, V.A, Pires, V.M.R, Flint, J.E, Pinheiro, B.A, Goyal, A, Prates, J.A.M, Izumi, A, Stalbrand, H, Kolenova, K, Topakas, E, Dodson, E.J, Bolam, D.N, Davies, G.J, Fontes, C.M.G.A, Gilbert, H.J. | | Deposit date: | 2009-02-04 | | Release date: | 2009-03-24 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The Active Site of a Carbohydrate Esterase Displays Divergent Catalytic and Noncatalytic Binding Functions.
Plos Biol., 7, 2009
|
|
5L0M
 
 | | hLRH-1 DNA Binding Domain - 12bp Oct4 promoter complex | | Descriptor: | DNA (5'-D(*CP*TP*AP*GP*CP*CP*TP*TP*GP*AP*CP*C)-3'), DNA (5'-D(*GP*GP*TP*CP*AP*AP*GP*GP*CP*TP*AP*G)-3'), GLYCEROL, ... | | Authors: | Tuntland, M.L, Ortlund, E.A. | | Deposit date: | 2016-07-27 | | Release date: | 2016-11-09 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.197 Å) | | Cite: | A Structural Investigation into Oct4 Regulation by Orphan Nuclear Receptors, Germ Cell Nuclear Factor (GCNF), and Liver Receptor Homolog-1 (LRH-1).
J. Mol. Biol., 428, 2016
|
|
3JZ1
 
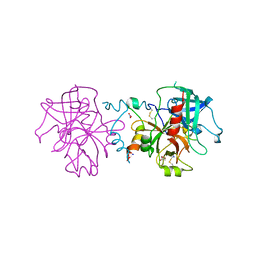 | | Crystal structure of human thrombin mutant N143P in E:Na+ form | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, NITRATE ION, ... | | Authors: | Niu, W, Chen, Z, Bush-Pelc, L.A, Bah, A, Gandhi, P.S, Di Cera, E. | | Deposit date: | 2009-09-22 | | Release date: | 2009-10-20 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Mutant N143P reveals how Na+ activates thrombin
J.Biol.Chem., 284, 2009
|
|
7JMT
 
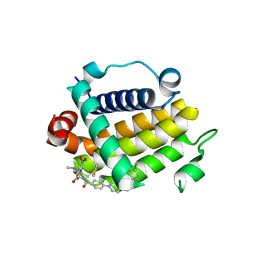 | | Crystal structure of schistosome BCL-2 bound to ABT-737 | | Descriptor: | 4-{4-[(4'-CHLOROBIPHENYL-2-YL)METHYL]PIPERAZIN-1-YL}-N-{[4-({(1R)-3-(DIMETHYLAMINO)-1-[(PHENYLTHIO)METHYL]PROPYL}AMINO)-3-NITROPHENYL]SULFONYL}BENZAMIDE, BCL-2 protein | | Authors: | Smith, N.A, Smith, B.J, Lee, E.F, Colman, P.M, Fairlie, W.D. | | Deposit date: | 2020-08-02 | | Release date: | 2021-02-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Optimization of Benzothiazole and Thiazole Hydrazones as Inhibitors of Schistosome BCL-2.
Acs Infect Dis., 7, 2021
|
|
7BNX
 
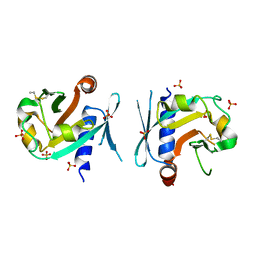 | | Archeal holliday junction resolvase from Thermus thermophilus phage 15-6 | | Descriptor: | Holliday junction resolvase, SULFATE ION | | Authors: | Hakansson, M, Ahlqvist, J, Linares Pasten, J.A, Jasilionis, A, Nordberg Karlsson, E, Al-Karadaghi, S. | | Deposit date: | 2021-01-22 | | Release date: | 2022-02-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.551 Å) | | Cite: | Crystal structure and initial characterization of a novel archaeal-like Holliday junction-resolving enzyme from Thermus thermophilus phage Tth15-6.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
6GXS
 
 | | Crystal structure of CV39L lectin from Chromobacterium violaceum at 1.8 A resolution | | Descriptor: | 1,2-ETHANEDIOL, CV39L lectin, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Sykorova, P, Novotna, J, Demo, G, Pompidor, G, Dubska, E, Komarek, J, Fujdiarova, E, Haronikova, L, Varrot, A, Imberty, A, Shilova, N, Bovin, N, Pokorna, M, Wimmerova, M. | | Deposit date: | 2018-06-27 | | Release date: | 2019-12-04 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Characterization of novel lectins from Burkholderia pseudomallei and Chromobacterium violaceum with seven-bladed beta-propeller fold.
Int.J.Biol.Macromol., 152, 2020
|
|
7S9Z
 
 | | Helicobacter Hepaticus CcsBA Closed Conformation | | Descriptor: | Cytochrome c biogenesis protein, HEME B/C, PHOSPHATIDYLETHANOLAMINE | | Authors: | Mendez, D.L, Lowder, E.P, Tillman, D.E, Sutherland, M.C, Collier, A.L, Rau, M.J, Fitzpatrick, J.A, Kranz, R.G. | | Deposit date: | 2021-09-21 | | Release date: | 2021-12-22 | | Last modified: | 2022-01-12 | | Method: | ELECTRON MICROSCOPY (4.14 Å) | | Cite: | Cryo-EM of CcsBA reveals the basis for cytochrome c biogenesis and heme transport.
Nat.Chem.Biol., 18, 2022
|
|
7BGS
 
 | | Archeal holliday junction resolvase from Thermus thermophilus phage 15-6 | | Descriptor: | Holliday junction resolvase, SULFATE ION | | Authors: | Hakansson, M, Ahlqvist, J, Linares Pasten, J.A, Jasilionis, A, Nordberg Karlsson, E, Al-Karadaghi, S. | | Deposit date: | 2021-01-08 | | Release date: | 2022-01-19 | | Last modified: | 2022-02-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure and initial characterization of a novel archaeal-like Holliday junction-resolving enzyme from Thermus thermophilus phage Tth15-6.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
4WPX
 
 | | Chaetomium theromophilum TREX2 CID domain complex | | Descriptor: | Cell division control protein 31-like protein, Putative SAC3 family protein, Putative uncharacterized protein | | Authors: | Valkov, E, Stewart, M. | | Deposit date: | 2014-10-21 | | Release date: | 2015-06-17 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.31 Å) | | Cite: | Structural Characterization of the Chaetomium thermophilum TREX-2 Complex and its Interaction with the mRNA Nuclear Export Factor Mex67:Mtr2.
Structure, 23, 2015
|
|
7S9Y
 
 | | Helicobacter Hepaticus CcsBA Open Conformation | | Descriptor: | Cytochrome c biogenesis protein, HEME B/C, PHOSPHATIDYLETHANOLAMINE | | Authors: | Mendez, D.L, Lowder, E.P, Tillman, D.E, Sutherland, M.C, Collier, A.L, Rau, M.J, Fitzpatrick, J.A, Kranz, R.G. | | Deposit date: | 2021-09-21 | | Release date: | 2021-12-22 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.56 Å) | | Cite: | Cryo-EM of CcsBA reveals the basis for cytochrome c biogenesis and heme transport.
Nat.Chem.Biol., 18, 2022
|
|
6Z9M
 
 | | Pseudoatomic model of the pre-fusion conformation of glycoprotein B of Herpes simplex virus 1 | | Descriptor: | Envelope glycoprotein B | | Authors: | Vollmer, B, Prazak, V, Vasishtan, D, Jefferys, E.E, Hernandez-Duran, A, Vallbracht, M, Klupp, B, Mettenleiter, T.C, Backovic, M, Rey, F.A, Topf, M, Gruenewald, K. | | Deposit date: | 2020-06-04 | | Release date: | 2020-10-07 | | Last modified: | 2020-10-14 | | Method: | ELECTRON MICROSCOPY (9.1 Å) | | Cite: | The prefusion structure of herpes simplex virus glycoprotein B.
Sci Adv, 6, 2020
|
|
2N7M
 
 | | NMR-SAXS/WAXS Structure of the core of the U4/U6 di-snRNA | | Descriptor: | RNA (92-MER) | | Authors: | Cornilescu, G, Didychuk, A.L, Rodgers, M.L, Michael, L.A, Burke, J.E, Montemayor, E.J, Hoskins, A.A, Butcher, S.E, Tonelli, M. | | Deposit date: | 2015-09-14 | | Release date: | 2015-12-30 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural Analysis of Multi-Helical RNAs by NMR-SAXS/WAXS: Application to the U4/U6 di-snRNA.
J.Mol.Biol., 428, 2016
|
|
5KRJ
 
 | | Crystal Structure of the ER-alpha Ligand-binding Domain (Y537S) in Complex with an a-naphthyl Substituted OBHS derivative | | Descriptor: | Estrogen receptor, NCOA2, naphthalen-1-yl (1~{S},2~{R},4~{S})-5,6-bis(4-hydroxyphenyl)-7-oxabicyclo[2.2.1]hept-5-ene-2-sulfonate | | Authors: | Nwachukwu, J.C, Srinivasan, S, Bruno, N.E, Nowak, J, Kojetin, D.J, Elemento, O, Katzenellenbogen, J.A, Nettles, K.W. | | Deposit date: | 2016-07-07 | | Release date: | 2017-01-18 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Systems Structural Biology Analysis of Ligand Effects on ER alpha Predicts Cellular Response to Environmental Estrogens and Anti-hormone Therapies.
Cell Chem Biol, 24, 2017
|
|
