7QYS
 
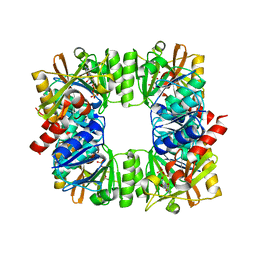 | | Crystal structure of RimK from Pseudomonas syringae DC3000 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Probable alpha-L-glutamate ligase | | Authors: | Thompson, C.M.A, Little, R.H, Stevenson, C.E.M, Lawson, D.M, Malone, J.G. | | Deposit date: | 2022-01-29 | | Release date: | 2022-10-05 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural insights into the mechanism of adaptive ribosomal modification by Pseudomonas RimK.
Proteins, 91, 2023
|
|
4Y7W
 
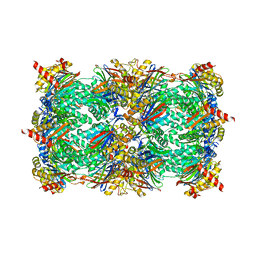 | | Yeast 20S proteasome in complex with Ac-LAE-ep | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Ac-LAE-ep, CHLORIDE ION, ... | | Authors: | Huber, E.M, Groll, M. | | Deposit date: | 2015-02-16 | | Release date: | 2015-06-17 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Systematic Analyses of Substrate Preferences of 20S Proteasomes Using Peptidic Epoxyketone Inhibitors.
J.Am.Chem.Soc., 137, 2015
|
|
4Y8J
 
 | | Yeast 20S proteasome in complex with Ac-LLL-ep | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Ac-PLL-ep, CHLORIDE ION, ... | | Authors: | Huber, E.M, Groll, M. | | Deposit date: | 2015-02-16 | | Release date: | 2015-06-17 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Systematic Analyses of Substrate Preferences of 20S Proteasomes Using Peptidic Epoxyketone Inhibitors.
J.Am.Chem.Soc., 137, 2015
|
|
6PKA
 
 | |
4YB6
 
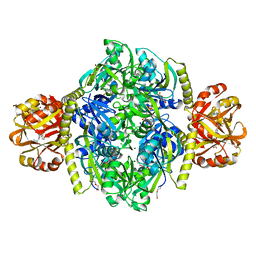 | | Adenosine triphosphate phosphoribosyltransferase from Campylobacter jejuni in complex with the inhibitors AMP and histidine | | Descriptor: | ADENOSINE MONOPHOSPHATE, ATP phosphoribosyltransferase, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Mittelstaedt, G, Moggre, G.-J, Parker, E.J. | | Deposit date: | 2015-02-18 | | Release date: | 2016-03-09 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Campylobacter jejuni adenosine triphosphate phosphoribosyltransferase is an active hexamer that is allosterically controlled by the twisting of a regulatory tail.
Protein Sci., 25, 2016
|
|
7QP2
 
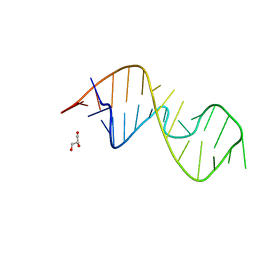 | | 1-deazaguanosine modified-RNA Sarcin Ricin Loop | | Descriptor: | GLYCEROL, RNA (27-MER) | | Authors: | Ennifar, E, Micura, R, Bereiter, R, Renard, E, Kreutz, C. | | Deposit date: | 2021-12-30 | | Release date: | 2022-07-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (0.9 Å) | | Cite: | 1-Deazaguanosine-Modified RNA: The Missing Piece for Functional RNA Atomic Mutagenesis.
J.Am.Chem.Soc., 144, 2022
|
|
8G1C
 
 | | Crystal structure of polyreactive 3B03 human Fab | | Descriptor: | Heavy chain of monoreactive 3B03 human Fab fragment, Light chain of monoreactive 3B03 human Fab fragment | | Authors: | Borowska, M.T, Adams, E.J. | | Deposit date: | 2023-02-01 | | Release date: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Biochemical and biophysical characterization of natural polyreactivity in antibodies.
Cell Rep, 42, 2023
|
|
8PE1
 
 | | Crystal structure of Gel4 in complex with Nanobody 4 | | Descriptor: | 1,3-beta-glucanosyltransferase, 2-acetamido-2-deoxy-beta-D-glucopyranose, Nanobody 4, ... | | Authors: | Macias-Leon, J, Redrado-Hernandez, S, Castro-Lopez, J, Sanz, A.B, Arias, M, Farkas, V, Vincke, C, Muyldermans, S, Pardo, J, Arroyo, J, Galvez, E, Hurtado-Guerrero, R. | | Deposit date: | 2023-06-13 | | Release date: | 2024-06-19 | | Last modified: | 2024-08-21 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Broad Protection against Invasive Fungal Disease from a Nanobody Targeting the Active Site of Fungal beta-1,3-Glucanosyltransferases.
Angew.Chem.Int.Ed.Engl., 63, 2024
|
|
8FAT
 
 | | Crystal structure of Ky224 Fab in complex with circumsporozoite protein NPDP peptide | | Descriptor: | Circumsporozoite protein NPDP peptide, Ky224 Antibody, heavy chain, ... | | Authors: | Kassardjian, A, Thai, E, Julien, J.P. | | Deposit date: | 2022-11-28 | | Release date: | 2023-11-01 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Molecular determinants of cross-reactivity and potency by VH3-33 antibodies against the Plasmodium falciparum circumsporozoite protein.
Cell Rep, 42, 2023
|
|
8FB5
 
 | | Crystal structure of Ky15.11-S100IK Fab in complex with circumsporozoite protein KQPA peptide | | Descriptor: | Circumsporozoite protein KQPA peptide, Ky15.11-SK Antibody, heavy chain, ... | | Authors: | Kang, R.W, Thai, E, Julien, J.P. | | Deposit date: | 2022-11-29 | | Release date: | 2023-11-01 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Molecular determinants of cross-reactivity and potency by VH3-33 antibodies against the Plasmodium falciparum circumsporozoite protein.
Cell Rep, 42, 2023
|
|
6YLJ
 
 | | Structure of D169A/E171A double mutant of chitinase Chit42 from Trichoderma harzianum complexed with chitinhexaose. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, Endochitinase 42, ... | | Authors: | Jimenez-Ortega, E, Sanz-Aparicio, J. | | Deposit date: | 2020-04-07 | | Release date: | 2021-10-20 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural inspection and protein motions modelling of a fungal glycoside hydrolase family 18 chitinase by crystallography depicts a dynamic enzymatic mechanism
Comput Struct Biotechnol J, 19, 2021
|
|
6YN4
 
 | | Structure of D169A/E171A double mutant of chitinase Chit42 from Trichoderma harzianum complexed with chitintetraose. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, ... | | Authors: | Jimenez-Ortega, E, Sanz-Aparicio, J. | | Deposit date: | 2020-04-10 | | Release date: | 2021-10-20 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Structural inspection and protein motions modelling of a fungal glycoside hydrolase family 18 chitinase by crystallography depicts a dynamic enzymatic mechanism
Comput Struct Biotechnol J, 19, 2021
|
|
8PE2
 
 | | Crystal structure of Gel4 in complex with Nanobody 3 | | Descriptor: | 1,3-beta-glucanosyltransferase, 2-acetamido-2-deoxy-beta-D-glucopyranose, Nanobody 3, ... | | Authors: | Macias-Leon, J, Redrado-Hernandez, S, Castro-Lopez, J, Sanz, A.B, Arias, M, Farkas, V, Vincke, C, Muyldermans, S, Pardo, J, Arroyo, J, Galvez, E, Hurtado-Guerrero, R. | | Deposit date: | 2023-06-13 | | Release date: | 2024-06-19 | | Last modified: | 2024-08-21 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Broad Protection against Invasive Fungal Disease from a Nanobody Targeting the Active Site of Fungal beta-1,3-Glucanosyltransferases.
Angew.Chem.Int.Ed.Engl., 63, 2024
|
|
4YA7
 
 | |
6UVG
 
 | | Crystal structure of BCL-XL bound to compound 13: (R)-2-(3-([1,1'-Biphenyl]-4-carbonyl)-3-(4-methylbenzyl)ureido)-3-(((3R,5R,7R)-adamantan-1-ylmethyl)sulfonyl)propanoic acid | | Descriptor: | (R)-2-(3-([1,1'-Biphenyl]-4-carbonyl)-3-(4-methylbenzyl)ureido)-3-(((3R,5R,7R)-adamantan-1-ylmethyl)sulfonyl)propanoic acid, 1,2-ETHANEDIOL, Bcl-2-like protein 1, ... | | Authors: | Roy, M.J, Lessene, G, Czabotar, P.E. | | Deposit date: | 2019-11-02 | | Release date: | 2021-05-05 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure-Guided Development of Potent Benzoylurea Inhibitors of BCL-X L and BCL-2.
J.Med.Chem., 64, 2021
|
|
8FTN
 
 | | E. coli ArnA dehydrogenase domain mutant - N492A | | Descriptor: | Bifunctional UDP-4-amino-4-deoxy-L-arabinose formyltransferase/UDP-glucuronic acid oxidase ArnA, SULFATE ION | | Authors: | Sousa, M.C, Mitchell, M.E. | | Deposit date: | 2023-01-12 | | Release date: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Targeting the Conformational Change in ArnA Dehydrogenase for Selective Inhibition of Polymyxin Resistance.
Biochemistry, 62, 2023
|
|
4YGA
 
 | |
6N2W
 
 | | The structure of Stable-5-Lipoxygenase bound to NDGA | | Descriptor: | 4-[(2R,3S)-3-[(3,4-DIHYDROXYPHENYL)METHYL]-2-METHYLBUTYL]BENZENE-1,2-DIOL, Arachidonate 5-lipoxygenase, FE (II) ION | | Authors: | Newcomer, M.E, Gilbert, N.C, Neau, D.B. | | Deposit date: | 2018-11-14 | | Release date: | 2020-05-13 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | Structural and mechanistic insights into 5-lipoxygenase inhibition by natural products.
Nat.Chem.Biol., 16, 2020
|
|
8UGZ
 
 | | Crystal structure of Shewanella benthica Group 1 truncated hemoglobin C51S C71S variant | | Descriptor: | CYANIDE ION, Group 1 truncated hemoglobin, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Schultz, T.D, Martinez, J.E, Siegler, M.A, Schlessman, J.L, Lecomte, J.T.J. | | Deposit date: | 2023-10-06 | | Release date: | 2024-04-03 | | Last modified: | 2024-07-17 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Heme d formation in a Shewanella benthica hemoglobin.
J.Inorg.Biochem., 259, 2024
|
|
4Y74
 
 | | Yeast 20S proteasome in complex with Ac-LAL-ep | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Ac-LAL-ep, CHLORIDE ION, ... | | Authors: | Groll, M, Huber, E.M. | | Deposit date: | 2015-02-13 | | Release date: | 2015-06-17 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Systematic Analyses of Substrate Preferences of 20S Proteasomes Using Peptidic Epoxyketone Inhibitors.
J.Am.Chem.Soc., 137, 2015
|
|
7QAN
 
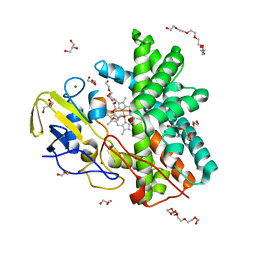 | | Cytochrome P450 Enzyme AbyV | | Descriptor: | 3,6,9,12,15,18-HEXAOXAICOSANE-1,20-DIOL, CHLORIDE ION, Cytochrome P450, ... | | Authors: | Parnell, A.E, Back, C.R, Race, P.R. | | Deposit date: | 2021-11-17 | | Release date: | 2022-11-30 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | The Role of Cytochrome P450 AbyV in the Final Stages of Abyssomicin C Biosynthesis.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
7OQZ
 
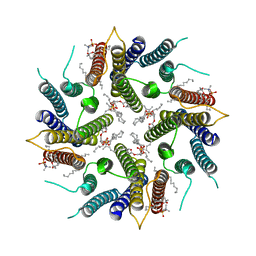 | | Cryo-EM structure of human TMEM45A | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, Transmembrane protein 45A | | Authors: | Grieben, M, Pike, A.C.W, Evans, A, Shrestha, L, Venkaya, S, Mukhopadhyay, S.M.M, Moreira, T, Chalk, R, MacLean, E.M, Marsden, B.D, Burgess-Brown, N.A, Bountra, C, Carpenter, E.P. | | Deposit date: | 2021-06-04 | | Release date: | 2021-06-16 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3.27 Å) | | Cite: | CryoEM structure of human TMEM45A
To Be Published
|
|
7TLL
 
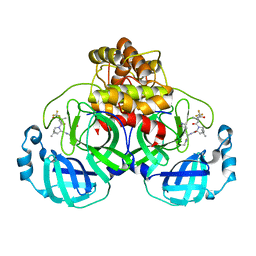 | | Structure of SARS-CoV-2 Mpro Omicron P132H in complex with Nirmatrelvir (PF-07321332) | | Descriptor: | (1R,2S,5S)-N-{(1E,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase nsp5 | | Authors: | Greasley, S.E, Ferre, R.A, Plotnikova, O, Liu, W, Stewart, A.E. | | Deposit date: | 2022-01-18 | | Release date: | 2022-01-26 | | Last modified: | 2022-06-15 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Structural basis for the in vitro efficacy of nirmatrelvir against SARS-CoV-2 variants.
J.Biol.Chem., 298, 2022
|
|
6ZFC
 
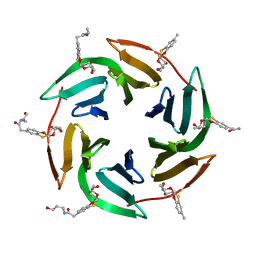 | | Fucose-binding lectin from Burkholderia ambifaria (BamBL) in complex with a fucosyl derivative | | Descriptor: | 2-[(2~{S},3~{S},4~{R},4~{a}~{S},10~{a}~{S})-2-methyl-3,4-bis(oxidanyl)-3,4,4~{a},10~{a}-tetrahydro-2~{H}-pyrano[2,3-b][1,4]benzoxathiin-7-yl]-~{N}-(3-oxidanylpropyl)ethanamide, bacterial lectin from Burkholderia ambifaria | | Authors: | Kuhaudomlarp, S, Gillon, E, Fragai, M, Cerofolini, L, Giuntini, S, Denis, M, Santarsia, S, Valori, C, Dondoni, A, Fallarini, S, Lombardi, G, Nativi, C, Imberty, A. | | Deposit date: | 2020-06-17 | | Release date: | 2020-10-28 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Fucosylated ubiquitin and orthogonally glycosylated mutant A28C: conceptually new ligands for Burkholderia ambifaria lectin (BambL).
Chem Sci, 11, 2020
|
|
6PB1
 
 | | Cryo-EM structure of Urocortin 1-bound Corticotropin-releasing factor 2 receptor in complex with Gs protein and Nb35 | | Descriptor: | CHOLESTEROL, Corticotropin-releasing factor receptor 2, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Ma, S, Shen, Q, Zhao, L.-H, Mao, C, Zhou, X.E, Shen, D.-D, de Waal, P.W, Bi, P, Li, C, Jiang, Y, Wang, M.-W, Sexton, P.M, Wootten, D, Melcher, K, Zhang, Y, Xu, H.E. | | Deposit date: | 2019-06-12 | | Release date: | 2020-02-12 | | Last modified: | 2020-02-19 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Molecular Basis for Hormone Recognition and Activation of Corticotropin-Releasing Factor Receptors.
Mol.Cell, 77, 2020
|
|
