6YOR
 
 | | Structure of the SARS-CoV-2 spike S1 protein in complex with CR3022 Fab | | Descriptor: | IgG H chain, IgG L chain, Spike glycoprotein | | Authors: | Huo, J, Zhao, Y, Ren, J, Zhou, D, Duyvesteyn, H.M.E, Carrique, L, Malinauskas, T, Ruza, R.R, Shah, P.N.M, Fry, E.E, Owens, R, Stuart, D.I. | | Deposit date: | 2020-04-15 | | Release date: | 2020-04-29 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Neutralization of SARS-CoV-2 by Destruction of the Prefusion Spike.
Cell Host Microbe, 28, 2020
|
|
6YE2
 
 | |
6HK0
 
 | | X-ray structure of a pentameric ligand gated ion channel from Erwinia chrysanthemi (ELIC) F16'S pore mutant (F247S) with alternate M4 conformation. | | Descriptor: | Cys-loop ligand-gated ion channel, DODECYL-BETA-D-MALTOSIDE | | Authors: | Nury, H, Spurny, R, Govaerts, C, Evans, G.L, Pardon, E, Steyaert, J, Ulens, C. | | Deposit date: | 2018-09-04 | | Release date: | 2019-10-09 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.45 Å) | | Cite: | A lipid site shapes the agonist response of a pentameric ligand-gated ion channel.
Nat.Chem.Biol., 15, 2019
|
|
6HCK
 
 | | The Transcriptional Regulator PrfA from Listeria Monocytogenes in complex with dipeptide Leu-Leu | | Descriptor: | LEUCINE, Listeriolysin regulatory protein, SODIUM ION | | Authors: | Grundstrom, C, Oelker, M, Krypotou, E, Scortti, M, Luisi, B.F, Vazquez-Boland, J, Sauer-Eriksson, A.E. | | Deposit date: | 2018-08-15 | | Release date: | 2019-02-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Control of Bacterial Virulence through the Peptide Signature of the Habitat.
Cell Rep, 26, 2019
|
|
6HR2
 
 | | Crystal structure of PROTAC 2 in complex with the bromodomain of human SMARCA4 and pVHL:ElonginC:ElonginB | | Descriptor: | (2~{S},4~{R})-~{N}-[[2-[2-[4-[[4-[3-azanyl-6-(2-hydroxyphenyl)pyridazin-4-yl]piperazin-1-yl]methyl]phenyl]ethoxy]-4-(4-methyl-1,3-thiazol-5-yl)phenyl]methyl]-1-[(2~{S})-2-[(1-fluoranylcyclopropyl)carbonylamino]-3,3-dimethyl-butanoyl]-4-oxidanyl-pyrrolidine-2-carboxamide, 1,2-ETHANEDIOL, DIMETHYL SULFOXIDE, ... | | Authors: | Roy, M, Bader, G, Diers, E, Trainor, N, Farnaby, W, Ciulli, A. | | Deposit date: | 2018-09-26 | | Release date: | 2019-06-12 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | BAF complex vulnerabilities in cancer demonstrated via structure-based PROTAC design.
Nat.Chem.Biol., 15, 2019
|
|
4X0P
 
 | |
6QGA
 
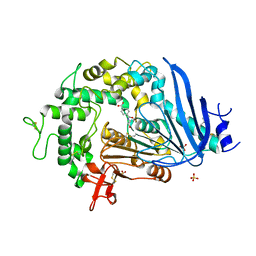 | | Crystal structure of Ideonella sakaiensis MHETase bound to the non-hydrolyzable ligand MHETA | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 4-(2-hydroxyethylcarbamoyl)benzoic acid, CALCIUM ION, ... | | Authors: | Palm, G.J, Reisky, L, Boettcher, D, Mueller, H, Michels, E.A.P, Walczak, C, Berndt, L, Weiss, M.S, Bornscheuer, U.T, Weber, G. | | Deposit date: | 2019-01-10 | | Release date: | 2019-04-03 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of the plastic-degrading Ideonella sakaiensis MHETase bound to a substrate.
Nat Commun, 10, 2019
|
|
6QGB
 
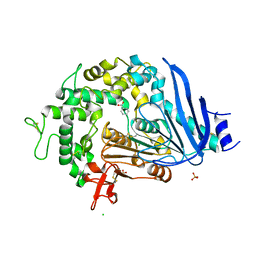 | | Crystal structure of Ideonella sakaiensis MHETase bound to benzoic acid | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, BENZOIC ACID, CALCIUM ION, ... | | Authors: | Palm, G.J, Reisky, L, Boettcher, D, Mueller, H, Michels, E.A.P, Walczak, C, Berndt, L, Weiss, M.S, Bornscheuer, U.T, Weber, G. | | Deposit date: | 2019-01-10 | | Release date: | 2019-04-03 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of the plastic-degrading Ideonella sakaiensis MHETase bound to a substrate.
Nat Commun, 10, 2019
|
|
9CHN
 
 | | P. vulgaris trimeric HigBA- operator 2 DNA | | Descriptor: | Antitoxin HigA, DNA (5'-D(*GP*TP*AP*TP*TP*AP*CP*AP*CP*AP*CP*CP*AP*TP*GP*TP*AP*AP*TP*AP*C)-3'), DNA (5'-D(*GP*TP*AP*TP*TP*AP*CP*AP*TP*GP*GP*TP*GP*TP*GP*TP*AP*AP*TP*AP*C)-3'), ... | | Authors: | Pavelich, I.J, Schureck, M.A, Hoffer, E.D, Dunham, C.M. | | Deposit date: | 2024-07-01 | | Release date: | 2025-07-02 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | P. vulgaris trimeric HigBA- operator 2 DNA
To Be Published
|
|
4YO5
 
 | | EAEC T6SS TssA-Cterminus | | Descriptor: | TssA | | Authors: | Durand, E, Zoued, A, Spinelli, S, Douzi, B, Brunet, Y.R, Bebeacua, C, Legrand, P, Journet, L, Mignot, T, Cambillau, C, Cascales, E. | | Deposit date: | 2015-03-11 | | Release date: | 2016-02-17 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (3.35 Å) | | Cite: | Priming and polymerization of a bacterial contractile tail structure.
Nature, 531, 2016
|
|
7ZJ4
 
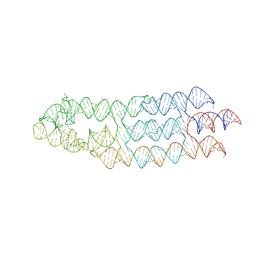 | | Ligand bound state of a brocolli-pepper aptamer FRET tile | | Descriptor: | 4-(3,5-difluoro-4-hydroxybenzyl)-1,2-dimethyl-1H-imidazol-5-ol, 4-[(~{Z})-1-cyano-2-[5-[2-hydroxyethyl(methyl)amino]thieno[3,2-b]thiophen-2-yl]ethenyl]benzenecarbonitrile, POTASSIUM ION, ... | | Authors: | McRae, E.K.S, Vallina, N.S, Hansen, B.K, Boussebayle, A, Andersen, E.S. | | Deposit date: | 2022-04-08 | | Release date: | 2023-04-19 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (4.43 Å) | | Cite: | Structure determination of Pepper-Broccoli FRET pair by RNA origami scaffolding
To Be Published
|
|
2NOQ
 
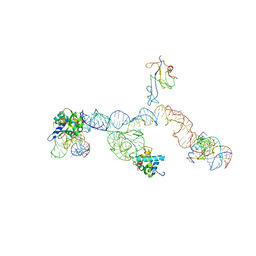 | | Structure of ribosome-bound cricket paralysis virus IRES RNA | | Descriptor: | 18S ribosomal RNA, 25S ribosomal RNA, 40S ribosomal protein S5, ... | | Authors: | Schuler, M, Connell, S.R, Lescoute, A, Giesebrecht, J, Dabrowski, M, Schroeer, B, Mielke, T, Penczek, P.A, Westhof, E, Spahn, C.M.T. | | Deposit date: | 2006-10-26 | | Release date: | 2006-11-21 | | Last modified: | 2023-12-27 | | Method: | ELECTRON MICROSCOPY (7.3 Å) | | Cite: | Structure of the ribosome-bound cricket paralysis virus IRES RNA.
Nat.Struct.Mol.Biol., 13, 2006
|
|
6F2J
 
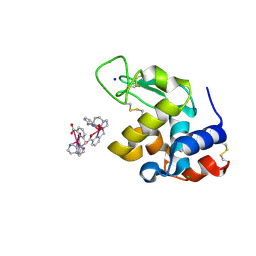 | | Crystal structure of Hen Egg-White Lysozyme co-crystallized in presence of 100 mM Tb-Xo4 and 100 mM sodium sulfate | | Descriptor: | CHLORIDE ION, Lysozyme C, SODIUM ION, ... | | Authors: | Engilberge, S, Riobe, F, Di Pietro, S, Girard, E, Dumont, E, Maury, O. | | Deposit date: | 2017-11-24 | | Release date: | 2018-10-03 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Unveiling the Binding Modes of the Crystallophore, a Terbium-based Nucleating and Phasing Molecular Agent for Protein Crystallography.
Chemistry, 24, 2018
|
|
7NPU
 
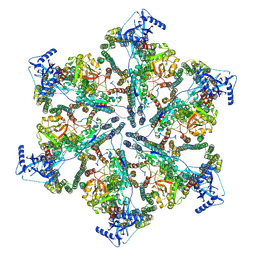 | | MycP5-free ESX-5 inner membrane complex, state I | | Descriptor: | ESX-5 secretion system ATPase EccB5, ESX-5 secretion system protein EccC5, ESX-5 secretion system protein EccD5 | | Authors: | Fahrenkamp, D, Bunduc, C.M, Wald, J, Ummels, R, Bitter, W, Houben, E.N.G, Marlovits, T.C. | | Deposit date: | 2021-02-28 | | Release date: | 2021-06-02 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (4.48 Å) | | Cite: | Structure and dynamics of a mycobacterial type VII secretion system.
Nature, 593, 2021
|
|
6EU2
 
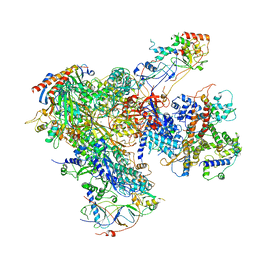 | | Apo RNA Polymerase III - open conformation (oPOL3) | | Descriptor: | DNA-directed RNA polymerase III subunit RPC1, DNA-directed RNA polymerase III subunit RPC10, DNA-directed RNA polymerase III subunit RPC2, ... | | Authors: | Abascal-Palacios, G, Ramsay, E.P, Beuron, F, Morris, E, Vannini, A. | | Deposit date: | 2017-10-27 | | Release date: | 2018-01-17 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural basis of RNA polymerase III transcription initiation.
Nature, 553, 2018
|
|
7NPR
 
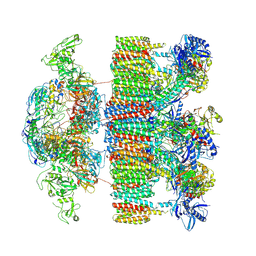 | | Structure of an intact ESX-5 inner membrane complex, Composite C3 model | | Descriptor: | ESX-5 secretion system ATPase EccB5, ESX-5 secretion system protein EccC5, ESX-5 secretion system protein EccD5, ... | | Authors: | Fahrenkamp, D, Bunduc, C.M, Wald, J, Ummels, R, Bitter, W, Houben, E.N.G, Marlovits, T.C. | | Deposit date: | 2021-02-28 | | Release date: | 2021-06-02 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.82 Å) | | Cite: | Structure and dynamics of a mycobacterial type VII secretion system.
Nature, 593, 2021
|
|
7NPV
 
 | | MycP5-free ESX-5 inner membrane complex, State II | | Descriptor: | ESX-5 secretion system ATPase EccB5, ESX-5 secretion system protein EccC5, ESX-5 secretion system protein EccD5 | | Authors: | Fahrenkamp, D, Bunduc, C.M, Wald, J, Ummels, R, Bitter, W, Houben, E.N.G, Marlovits, T.C. | | Deposit date: | 2021-02-28 | | Release date: | 2021-06-02 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (6.66 Å) | | Cite: | Structure and dynamics of a mycobacterial type VII secretion system.
Nature, 593, 2021
|
|
8FJT
 
 | | Human mitochondrial serine hydroxymethyltransferase (SHMT2) in complex with PLP, glycine and AGF362 inhibitor | | Descriptor: | GLYCINE, N-GLYCINE-[3-HYDROXY-2-METHYL-5-PHOSPHONOOXYMETHYL-PYRIDIN-4-YL-METHANE], N-{4-[4-(2-amino-4-oxo-3,4-dihydro-5H-pyrrolo[3,2-d]pyrimidin-5-yl)butyl]-3-fluorothiophene-2-carbonyl}-L-glutamic acid, ... | | Authors: | Katinas, J.M, Dann III, C.E. | | Deposit date: | 2022-12-20 | | Release date: | 2023-09-06 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.47 Å) | | Cite: | Structure-Based Design of Transport-Specific Multitargeted One-Carbon Metabolism Inhibitors in Cytosol and Mitochondria.
J.Med.Chem., 66, 2023
|
|
7NL2
 
 | | Structure of Xyn11 from Pseudothermotoga thermarum | | Descriptor: | 1-methylethyl 1-thio-beta-D-galactopyranoside, Beta-xylanase, GLYCEROL | | Authors: | Jimenez-Ortega, E, Sanz-Aparicio, J. | | Deposit date: | 2021-02-22 | | Release date: | 2021-06-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Phylogenetic, functional and structural characterization of a GH10 xylanase active at extreme conditions of temperature and alkalinity
Comput Struct Biotechnol J, 19, 2021
|
|
6OTS
 
 | | Rat ERK2 E320K | | Descriptor: | Mitogen-activated protein kinase 1 | | Authors: | Taylor, C.A, Cormier, K.W, Juang, Y.-C, Goldsmith, E.J, Cobb, M.H. | | Deposit date: | 2019-05-03 | | Release date: | 2019-07-10 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Functional divergence caused by mutations in an energetic hotspot in ERK2.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
8IU0
 
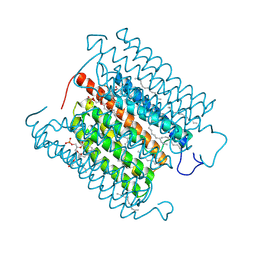 | | Cryo-EM structure of the potassium-selective channelrhodopsin HcKCR1 H225F mutant in lipid nanodisc | | Descriptor: | (7R,17E,20E)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSA-17,20-DIEN-1-AMINIUM 4-OXIDE, HcKCR1, PALMITIC ACID, ... | | Authors: | Tajima, S, Kim, Y, Nakamura, S, Yamashita, K, Fukuda, M, Deisseroth, K, Kato, H.E. | | Deposit date: | 2023-03-23 | | Release date: | 2023-09-06 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (2.66 Å) | | Cite: | Structural basis for ion selectivity in potassium-selective channelrhodopsins.
Cell, 186, 2023
|
|
8FJU
 
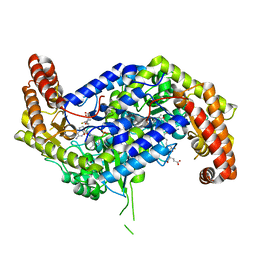 | | Human mitochondrial serine hydroxymethyltransferase (SHMT2) in complex with PLP, glycine and AGF347 inhibitor | | Descriptor: | N-GLYCINE-[3-HYDROXY-2-METHYL-5-PHOSPHONOOXYMETHYL-PYRIDIN-4-YL-METHANE], N-{4-[4-(2-amino-4-oxo-3,4-dihydro-5H-pyrrolo[3,2-d]pyrimidin-5-yl)butyl]-2-fluorobenzoyl}-L-glutamic acid, Serine hydroxymethyltransferase, ... | | Authors: | Katinas, J.M, Dann III, C.E. | | Deposit date: | 2022-12-20 | | Release date: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Structure-Based Design of Transport-Specific Multitargeted One-Carbon Metabolism Inhibitors in Cytosol and Mitochondria.
J.Med.Chem., 66, 2023
|
|
6ZIK
 
 | | bovine ATP synthase rotor domain, state 3 | | Descriptor: | ATP synthase F(0) complex subunit C2, mitochondrial, ATP synthase subunit delta, ... | | Authors: | Spikes, T, Montgomery, M.G, Walker, J.E. | | Deposit date: | 2020-06-26 | | Release date: | 2020-09-09 | | Last modified: | 2025-04-09 | | Method: | ELECTRON MICROSCOPY (3.66 Å) | | Cite: | Structure of the dimeric ATP synthase from bovine mitochondria.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6ET9
 
 | | Structure of the acetoacetyl-CoA-thiolase/HMG-CoA-synthase complex from Methanothermococcus thermolithotrophicus at 2.75 A | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Acetyl-CoA acetyltransferase thiolase, CHLORIDE ION, ... | | Authors: | Engilberge, S, Voegeli, B, Girard, E, Riobe, F, Maury, O, Erb, T.J, Shima, S, Wagner, T. | | Deposit date: | 2017-10-25 | | Release date: | 2018-03-14 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Archaeal acetoacetyl-CoA thiolase/HMG-CoA synthase complex channels the intermediate via a fused CoA-binding site.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6WPS
 
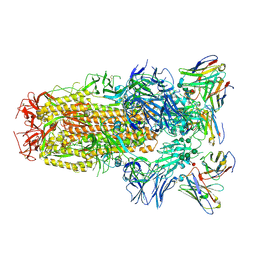 | | Structure of the SARS-CoV-2 spike glycoprotein in complex with the S309 neutralizing antibody Fab fragment | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, S309 neutralizing antibody heavy chain, ... | | Authors: | Pinto, D, Park, Y.J, Beltramello, M, Walls, A.C, Tortorici, M.A, Bianchi, S, Jaconi, S, Culap, K, Zatta, F, De Marco, A, Peter, A, Guarino, B, Spreafico, R, Cameroni, E, Case, J.B, Chen, R.E, Havenar-Daughton, C, Snell, G, Virgin, H.W, Lanzavecchia, A, Diamond, M.S, Fink, K, Veesler, D, Corti, D, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2020-04-27 | | Release date: | 2020-05-27 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Cross-neutralization of SARS-CoV-2 by a human monoclonal SARS-CoV antibody.
Nature, 583, 2020
|
|
