4WOH
 
 | | Structure of of human dual-specificity phosphatase 22 (E24A/K28A/K30A/C88S) complexed with 4-nitrophenolphosphate | | Descriptor: | 1,2-ETHANEDIOL, 4-NITROPHENYL PHOSPHATE, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Lountos, G.T, Cherry, S, Tropea, J.E, Waugh, D.S. | | Deposit date: | 2014-10-15 | | Release date: | 2015-02-18 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | Structural analysis of human dual-specificity phosphatase 22 complexed with a phosphotyrosine-like substrate.
Acta Crystallogr.,Sect.F, 71, 2015
|
|
4WUQ
 
 | | Crystal structure of human carbonic anhydrase isozyme I with 2,3,5,6-Tetrafluoro-4-piperidin-1-ylbenzenesulfonamide | | Descriptor: | 2,3,5,6-tetrafluoro-4-(piperidin-1-yl)benzenesulfonamide, Carbonic anhydrase 1, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Manakova, E, Smirnov, A, Grazulis, S. | | Deposit date: | 2014-11-03 | | Release date: | 2015-07-01 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Intrinsic thermodynamics of 4-substituted-2,3,5,6-tetrafluorobenzenesulfonamide binding to carbonic anhydrases by isothermal titration calorimetry.
Biophys.Chem., 205, 2015
|
|
6BN8
 
 | | Crystal structure of DDB1-CRBN-BRD4(BD1) complex bound to dBET55 PROTAC. | | Descriptor: | Bromodomain-containing protein 4, DNA damage-binding protein 1,DNA damage-binding protein 1, Protein cereblon, ... | | Authors: | Nowak, R.P, DeAngelo, S.L, Buckley, D, Bradner, J.E, Fischer, E.S. | | Deposit date: | 2017-11-16 | | Release date: | 2018-06-06 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.990035 Å) | | Cite: | Plasticity in binding confers selectivity in ligand-induced protein degradation.
Nat. Chem. Biol., 14, 2018
|
|
6SCI
 
 | | Structure of AdhE form 1 | | Descriptor: | Aldehyde-alcohol dehydrogenase, FE (III) ION | | Authors: | Lovering, A.L, Bragginton, E. | | Deposit date: | 2019-07-24 | | Release date: | 2020-08-26 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | High-resolution structure of the alcohol dehydrogenase domain of the bifunctional bacterial enzyme AdhE.
Acta Crystallogr.,Sect.F, 76, 2020
|
|
4WVI
 
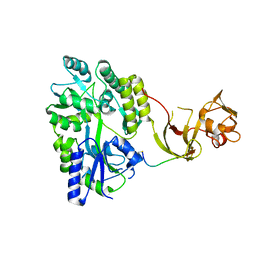 | | Crystal structure of the Type-I signal peptidase from Staphylococcus aureus (SpsB) in complex with a substrate peptide (pep2). | | Descriptor: | Maltose-binding periplasmic protein,Signal peptidase IB, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, substrate peptide (pep2) | | Authors: | Young, P.G, Ting, Y.T, Baker, E.N. | | Deposit date: | 2014-11-05 | | Release date: | 2015-09-23 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Peptide binding to a bacterial signal peptidase visualized by peptide tethering and carrier-driven crystallization.
IUCrJ, 3, 2016
|
|
6GM1
 
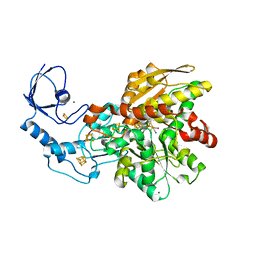 | | [FeFe]-hydrogenase CpI from Clostridium pasteurianum, variant E282A | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, IRON/SULFUR CLUSTER, Iron hydrogenase 1, ... | | Authors: | Duan, J, Esselborn, J, Hofmann, E, Winkler, M, Happe, T. | | Deposit date: | 2018-05-24 | | Release date: | 2018-11-07 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystallographic and spectroscopic assignment of the proton transfer pathway in [FeFe]-hydrogenases.
Nat Commun, 9, 2018
|
|
4WUI
 
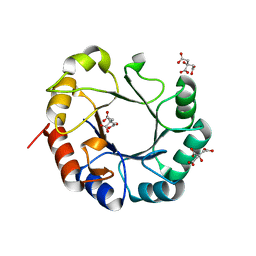 | | Crystal structure of TrpF from Jonesia denitrificans | | Descriptor: | CITRIC ACID, N-(5'-phosphoribosyl)anthranilate isomerase | | Authors: | Michalska, K, Verduzco-Castro, E.A, Endres, M, Barona-Gomez, F, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2014-10-31 | | Release date: | 2014-11-26 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.09 Å) | | Cite: | Co-occurrence of analogous enzymes determines evolution of a novel ( beta alpha )8-isomerase sub-family after non-conserved mutations in flexible loop.
Biochem. J., 473, 2016
|
|
4WUO
 
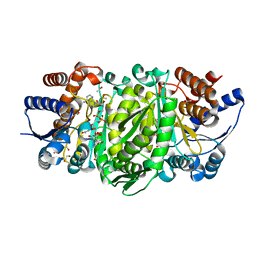 | | Structure of the E270A Mutant Isopropylmalate dehydrogenase from Thermus thermophilus in complex with IPM, Mn and NADH | | Descriptor: | 3-ISOPROPYLMALIC ACID, 3-isopropylmalate dehydrogenase, ETHANOL, ... | | Authors: | Pallo, A, Graczer, E, Olah, J, Szimler, T, Konarev, P.V, Svergun, D.I, Merli, A, Zavodszky, P, Vas, M, Weiss, M.S. | | Deposit date: | 2014-11-03 | | Release date: | 2014-11-12 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Glutamate 270 plays an essential role in K(+)-activation and domain closure of Thermus thermophilus isopropylmalate dehydrogenase.
Febs Lett., 589, 2015
|
|
6TW3
 
 | | HumRadA2 in complex with Naphthyl-HPA fragment-peptide chimera | | Descriptor: | (2~{S})-1-[(2~{S})-2-[(3-azanylnaphthalen-2-yl)carbonylamino]-3-(1~{H}-imidazol-4-yl)propanoyl]-~{N}-[(2~{S})-1-azanyl-1-oxidanylidene-propan-2-yl]pyrrolidine-2-carboxamide, DNA repair and recombination protein RadA, PHOSPHATE ION | | Authors: | Marsh, M.E, Fischer, G, Scott, D.E, Coyne, A.G, Skidmore, J, Abell, C, Hyvonen, M. | | Deposit date: | 2020-01-12 | | Release date: | 2021-01-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.352 Å) | | Cite: | A small-molecule inhibitor of the BRCA2-RAD51 interaction modulates RAD51 assembly and potentiates DNA damage-induced cell death.
Cell Chem Biol, 28, 2021
|
|
6F7C
 
 | | TUBULIN-Compound 12 complex | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 3,4,5-trimethoxy-~{N}-[(~{E})-naphthalen-1-ylmethylideneamino]benzamide, CALCIUM ION, ... | | Authors: | Muehlethaler, T, Prota, A.E, Steinmetz, M.O. | | Deposit date: | 2017-12-08 | | Release date: | 2018-12-19 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.002 Å) | | Cite: | Structural Basis of Colchicine-Site targeting Acylhydrazones active against Multidrug-Resistant Acute Lymphoblastic Leukemia.
Iscience, 21, 2019
|
|
1UVQ
 
 | | Crystal structure of HLA-DQ0602 in complex with a hypocretin peptide | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETIC ACID, GLYCINE, ... | | Authors: | Siebold, C, Hansen, B.E, Wyer, J.R, Harlos, K, Esnouf, R.E, Svejgaard, A, Bell, J.I, Strominger, J.L, Jones, E.Y, Fugger, L. | | Deposit date: | 2004-01-22 | | Release date: | 2004-02-05 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of Hla-Dq0602 that Protects Against Type 1 Diabetes and Confers Strong Susceptibility to Narcolepsy
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
8B5Y
 
 | | SHP2 in complex with allosteric imidazopyrazine inhibitor | | Descriptor: | (1~{S})-1'-[5-[2-(trifluoromethyl)pyridin-3-yl]sulfanyl-3~{H}-imidazo[4,5-b]pyrazin-2-yl]spiro[1,3-dihydroindene-2,4'-piperidine]-1-amine, FORMIC ACID, Tyrosine-protein phosphatase non-receptor type 11 | | Authors: | Torrente, E, Petrocchi, A. | | Deposit date: | 2022-09-25 | | Release date: | 2023-01-18 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Discovery of a Novel Series of Imidazopyrazine Derivatives as Potent SHP2 Allosteric Inhibitors.
Acs Med.Chem.Lett., 14, 2023
|
|
7TUJ
 
 | | NMR solution structure of the phosphorylated MUS81-binding region from human SLX4 | | Descriptor: | Structure-specific endonuclease subunit SLX4 | | Authors: | Payliss, B.J, Reichheld, S.E, Lemak, A, Arrowsmith, C.H, Sharpe, S, Wyatt, H.D.M. | | Deposit date: | 2022-02-02 | | Release date: | 2022-10-19 | | Last modified: | 2024-11-13 | | Method: | SOLUTION NMR | | Cite: | Phosphorylation of the DNA repair scaffold SLX4 drives folding of the SAP domain and activation of the MUS81-EME1 endonuclease.
Cell Rep, 41, 2022
|
|
4WHM
 
 | | Crystal structure of UDP-glucose: anthocyanidin 3-O-glucosyltransferase in complex with UDP | | Descriptor: | ACETATE ION, GLYCEROL, UDP-glucose:anthocyanidin 3-O-glucosyltransferase, ... | | Authors: | Hiromoto, T, Honjo, E, Tamada, T, Kuroki, R. | | Deposit date: | 2014-09-23 | | Release date: | 2015-01-21 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.851 Å) | | Cite: | Structural basis for acceptor-substrate recognition of UDP-glucose: anthocyanidin 3-O-glucosyltransferase from Clitoria ternatea
Protein Sci., 24, 2015
|
|
6OU2
 
 | |
7TD6
 
 | | aRML prion fibril | | Descriptor: | Major prion protein | | Authors: | Hoyt, F, Standke, H.G, Artikis, E, Caughey, B, Kraus, A. | | Deposit date: | 2021-12-30 | | Release date: | 2022-07-20 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Cryo-EM structure of anchorless RML prion reveals variations in shared motifs between distinct strains.
Nat Commun, 13, 2022
|
|
6MLX
 
 | | Crystal structure of T. pallidum Leucine Rich Repeat protein (TpLRR) | | Descriptor: | Leucine-rich repeat protein TpLRR | | Authors: | Ramaswamy, R, Loveless, B.C, Houston, S, Cameron, C.E, Boulanger, M.J. | | Deposit date: | 2018-09-28 | | Release date: | 2019-07-17 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural characterization of Treponema pallidum Tp0225 reveals an unexpected leucine-rich repeat architecture.
Acta Crystallogr.,Sect.F, 75, 2019
|
|
5FAN
 
 | | Crystal structure of PvHCT in complex with p-coumaroyl-CoA and protocatechuate | | Descriptor: | 3,4-DIHYDROXYBENZOIC ACID, Hydroxycinnamoyl-CoA shikimate/quinate hydroxycinnamoyltransferase 2, p-coumaroyl-CoA | | Authors: | Pereira, J.H, Moriarty, N.W, Eudes, A, Yogiswara, S, Wang, G, Benites, V.T, Baidoo, E.E.K, Lee, T.S, Keasling, J.D, Loque, D, Adams, P.D. | | Deposit date: | 2015-12-11 | | Release date: | 2016-02-24 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.901 Å) | | Cite: | Exploiting the Substrate Promiscuity of Hydroxycinnamoyl-CoA:Shikimate Hydroxycinnamoyl Transferase to Reduce Lignin.
Plant Cell.Physiol., 57, 2016
|
|
7U0S
 
 | | Crystal Structure of FK506-binding protein 1A from Aspergillus fumigatus Bound to Ascomycin | | Descriptor: | (3S,4R,5S,8R,9E,12S,14S,15R,16S,18R,19R,22R,26aS)-8-ethyl-5,19-dihydroxy-3-{(1E)-1-[(1R,3R,4R)-4-hydroxy-3-methoxycyclohexyl]prop-1-en-2-yl}-14,16-dimethoxy-4,10,12,18-tetramethyl-5,6,8,11,12,13,14,15,16,17,18,19,24,25,26,26a-hexadecahydro-3H-15,19-epoxypyrido[2,1-c][1,4]oxazacyclotricosine-1,7,20,21(4H,23H)-tetrone, ACETATE ION, FK506-binding protein 1A, ... | | Authors: | DeBouver, N.D, Fox III, D, Hoy, M.J, Heitman, J, Lorimer, D.D, Horanyi, P.S, Edwards, T.E, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2022-02-18 | | Release date: | 2022-07-27 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure-Guided Synthesis of FK506 and FK520 Analogs with Increased Selectivity Exhibit In Vivo Therapeutic Efficacy against Cryptococcus.
Mbio, 13, 2022
|
|
6FHN
 
 | | Structural dynamics and catalytic properties of a multi-modular xanthanase (Pt derivative) | | Descriptor: | 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Moroz, O.V, Jensen, P.F, McDonald, S.P, McGregor, N, Blagova, E, Comamala, G, Segura, D.R, Anderson, L, Vasu, S.M, Rao, V.P, Giger, L, Monrad, R.N, Svendsen, A, Nielsen, J.E, Henrissat, B, Davies, G.J, Brumer, H, Rand, K, Wilson, K.S. | | Deposit date: | 2018-01-15 | | Release date: | 2018-08-29 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Dynamics and Catalytic Properties of a Multimodular Xanthanase
Acs Catalysis, 2018
|
|
6BN9
 
 | | Crystal structure of DDB1-CRBN-BRD4(BD1) complex bound to dBET70 PROTAC | | Descriptor: | Bromodomain-containing protein 4, DNA damage-binding protein 1,DNA damage-binding protein 1, Protein cereblon, ... | | Authors: | Nowak, R.P, DeAngelo, S.L, Buckley, D, Bradner, J.E, Fischer, E.S. | | Deposit date: | 2017-11-16 | | Release date: | 2018-05-30 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (4.382 Å) | | Cite: | Plasticity in binding confers selectivity in ligand-induced protein degradation.
Nat. Chem. Biol., 14, 2018
|
|
4WW6
 
 | | Crystal structure of human carbonic anhydrase isozyme II with 2,3,5,6-Tetrafluoro-4-(propylthio)benzenesulfonamide | | Descriptor: | 2,3,5,6-tetrafluoro-4-(propylsulfanyl)benzenesulfonamide, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, BICINE, ... | | Authors: | Smirnov, A, Manakova, E, Grazulis, S. | | Deposit date: | 2014-11-10 | | Release date: | 2015-07-01 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.06 Å) | | Cite: | Intrinsic thermodynamics of 4-substituted-2,3,5,6-tetrafluorobenzenesulfonamide binding to carbonic anhydrases by isothermal titration calorimetry.
Biophys.Chem., 205, 2015
|
|
6TMQ
 
 | | Rapid optimisation of fragments and hits to lead compounds from screening of crude reaction mixtures | | Descriptor: | DIMETHYL SULFOXIDE, N-(2-AMINOETHYL)-2-{3-CHLORO-4-[(4-ISOPROPYLBENZYL)OXY]PHENYL} ACETAMIDE, [2,4-bis(oxidanyl)phenyl]-[(1~{S})-6,7-dimethoxy-1-pyridin-3-yl-3,4-dihydro-1~{H}-isoquinolin-2-yl]methanone, ... | | Authors: | Baker, L.M, Aimon, A, Murray, J.B, Surgenor, A.E, Matassova, N, Roughley, S.D, von Delft, F, Hubbard, R.E. | | Deposit date: | 2019-12-05 | | Release date: | 2020-10-14 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.112 Å) | | Cite: | Rapid optimisation of fragments and hits to lead compounds from screening of crude reaction mixtures
Commun Chem, 2020
|
|
7U0U
 
 | | Crystal Structure of a Aspergillus fumigatus Calcineurin A - Calcineurin B fusion bound to FKBP12 and FK-506 | | Descriptor: | 8-DEETHYL-8-[BUT-3-ENYL]-ASCOMYCIN, CALCIUM ION, PHOSPHATE ION, ... | | Authors: | Fox III, D, Abendroth, J, DeBouver, N.D, Hoy, M.J, Heitman, J, Lorimer, D.D, Horanyi, P.S, Edwards, T.E, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2022-02-18 | | Release date: | 2022-08-03 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure-Guided Synthesis of FK506 and FK520 Analogs with Increased Selectivity Exhibit In Vivo Therapeutic Efficacy against Cryptococcus.
Mbio, 13, 2022
|
|
5FAS
 
 | | OXA-48 in complex with FPI-1523 | | Descriptor: | Beta-lactamase, CADMIUM ION, CHLORIDE ION, ... | | Authors: | King, A.M, King, D.T, French, S, Brouillette, E, Asli, A, Alexander, A.N, Vuckovic, M, Maiti, S.N, Parr, T.R, Brown, E.D, Malouin, F, Strynadka, N.C.J, Wright, G.D. | | Deposit date: | 2015-12-11 | | Release date: | 2016-01-20 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Structural and Kinetic Characterization of Diazabicyclooctanes as Dual Inhibitors of Both Serine-beta-Lactamases and Penicillin-Binding Proteins.
Acs Chem.Biol., 11, 2016
|
|
