7S3P
 
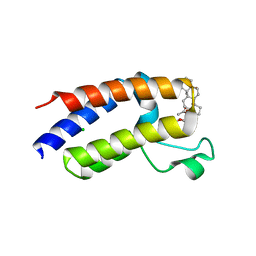 | | BD2 domain of human BRD3 bound to Physachenolide C | | Descriptor: | Bromodomain-containing protein 3, CHLORIDE ION, Physachenolide C | | Authors: | Horton, N.C, Chapman, E, Sivinski, J, Zerio, C, Ghadirian, N. | | Deposit date: | 2021-09-07 | | Release date: | 2023-01-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.89 Å) | | Cite: | Physachenolide C is a Potent, Selective BET Inhibitor.
J.Med.Chem., 66, 2023
|
|
8HK2
 
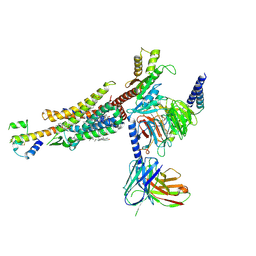 | | C3aR-Gi-C3a protein complex | | Descriptor: | C3a anaphylatoxin, C3a anaphylatoxin chemotactic receptor, CHOLESTEROL, ... | | Authors: | Wang, Y, Liu, W, Xu, Y, Zhuang, Y, Xu, H.E. | | Deposit date: | 2022-11-24 | | Release date: | 2023-05-10 | | Last modified: | 2023-11-08 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Revealing the signaling of complement receptors C3aR and C5aR1 by anaphylatoxins.
Nat.Chem.Biol., 19, 2023
|
|
2RVH
 
 | | NMR structure of eIF1 | | Descriptor: | Eukaryotic translation initiation factor eIF-1 | | Authors: | Nagata, T, Obayashi, E, Asano, K. | | Deposit date: | 2015-10-16 | | Release date: | 2016-10-26 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Molecular Landscape of the Ribosome Pre-initiation Complex during mRNA Scanning: Structural Role for eIF3c and Its Control by eIF5
Cell Rep, 18, 2017
|
|
3MMC
 
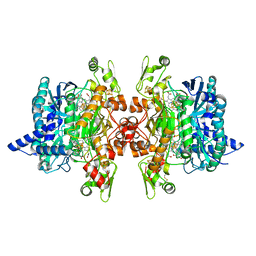 | | Structure of the dissimilatory sulfite reductase from Archaeoglobus fulgidus | | Descriptor: | GLYCEROL, IRON/SULFUR CLUSTER, SIROHEME, ... | | Authors: | Schiffer, A, Parey, K, Warkentin, E, Diederichs, K, Huber, H, Stetter, K.O, Kroneck, P.M.H, Ermler, U. | | Deposit date: | 2010-04-19 | | Release date: | 2010-05-12 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Structure of the dissimilatory sulfite reductase from the hyperthermophilic archaeon Archaeoglobus fulgidus.
J.Mol.Biol., 379, 2008
|
|
1AXV
 
 | | SOLUTION NMR STRUCTURE OF THE [BP]DA ADDUCT OPPOSITE DT IN A DNA DUPLEX, 6 STRUCTURES | | Descriptor: | 1,2,3-TRIHYDROXY-1,2,3,4-TETRAHYDROBENZO[A]PYRENE, DNA DUPLEX D(CTCTC-[BP]A-CTTCC)D(GGAAGTGAGAG) | | Authors: | Mao, B, Gu, Z, Gorin, A.A, Chen, J, Hingerty, B.E, Amid, S, Broyde, S, Geacintov, N.E, Patel, D.J. | | Deposit date: | 1997-10-21 | | Release date: | 1998-07-01 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the (+)-cis-anti-benzo[a]pyrene-dA ([BP]dA) adduct opposite dT in a DNA duplex.
Biochemistry, 38, 1999
|
|
5YTL
 
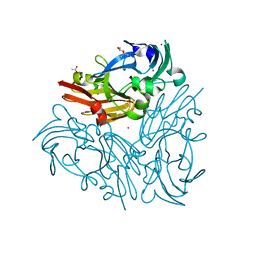 | | Crystal structure of Geobacillus thermodenitrificans copper-containing nitrite reductase determined with an anaerobically manipulated crystal | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, ACETIC ACID, ... | | Authors: | Fukuda, Y, Matsusaki, T, Tse, K.M, Mizohata, E, Murphy, M.E.P, Inoue, T. | | Deposit date: | 2017-11-19 | | Release date: | 2018-08-22 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.401 Å) | | Cite: | Crystallographic study of dioxygen chemistry in a copper-containing nitrite reductase from Geobacillus thermodenitrificans.
Acta Crystallogr D Struct Biol, 74, 2018
|
|
1BI3
 
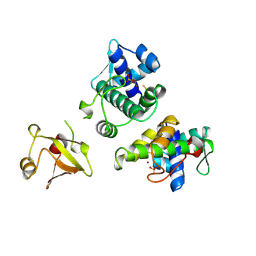 | | STRUCTURE OF APO-AND HOLO-DIPHTHERIA TOXIN REPRESSOR | | Descriptor: | DIPHTHERIA TOXIN REPRESSOR, SULFATE ION, ZINC ION | | Authors: | Pohl, E, Hol, W.G.J. | | Deposit date: | 1998-06-21 | | Release date: | 1999-06-22 | | Last modified: | 2022-12-21 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Motion of the DNA-binding domain with respect to the core of the diphtheria toxin repressor (DtxR) revealed in the crystal structures of apo- and holo-DtxR.
J.Biol.Chem., 273, 1998
|
|
6H78
 
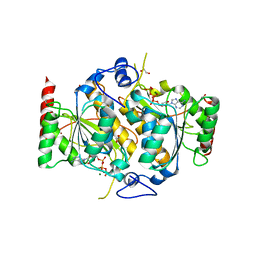 | | E1 enzyme for ubiquitin like protein activation. | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5'-TRIPHOSPHATE, CHLORIDE ION, ... | | Authors: | Soudah, N, Padala, P, Hassouna, F, Mashahreh, B, Lebedev, A.A, Isupov, M.N, Cohen-Kfir, E, Wiener, R. | | Deposit date: | 2018-07-30 | | Release date: | 2018-10-31 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | An N-Terminal Extension to UBA5 Adenylation Domain Boosts UFM1 Activation: Isoform-Specific Differences in Ubiquitin-like Protein Activation.
J.Mol.Biol., 431, 2019
|
|
3MQ1
 
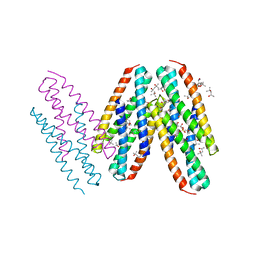 | | Crystal Structure of Dust Mite Allergen Der p 5 | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, Mite allergen Der p 5, ... | | Authors: | Mueller, G.A, Gosavi, R.A, Krahn, J.M, Edwards, L.L, Cuneo, M.J, Glesner, J, Pomes, A, Chapman, M.D, London, R.E, Pedersen, L.C. | | Deposit date: | 2010-04-27 | | Release date: | 2010-06-02 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Der p 5 crystal structure provides insight into the group 5 dust mite allergens.
J.Biol.Chem., 285, 2010
|
|
3M2L
 
 | |
7AIR
 
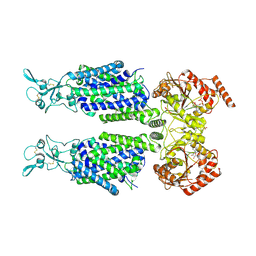 | | Structure of Human Potassium Chloride Transporter KCC1 in NaCl (Subclass 2) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Ebenhoch, R, Chi, G, Man, H, Wang, D, McKinley, G, Mukhopadhyay, S.M.M, MacLean, E.M, Chalk, R, Moreau, C, Snee, M, Bohstedt, T, Singh, N.K, Abrusci, P, Liko, I, Tehan, B.G, Almeida, F.G, Arrowsmith, C.H, Tang, H, Robinson, C.V, Bountra, C, Edwards, A.M, Marsden, B.D, Burgess-Brown, N.A, Duerr, K.L, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-09-28 | | Release date: | 2021-06-02 | | Last modified: | 2021-07-28 | | Method: | ELECTRON MICROSCOPY (3.66 Å) | | Cite: | Phospho-regulation, nucleotide binding and ion access control in potassium-chloride cotransporters.
Embo J., 40, 2021
|
|
5Y5J
 
 | | Time-resolved SFX structure of cytochrome P450nor: dark-2 data in the presence of NADH (resting state) | | Descriptor: | GLYCEROL, NADP nitrous oxide-forming nitric oxide reductase, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Tosha, T, Nomura, T, Nishida, T, Saeki, N, Okubayashi, K, Yamagiwa, R, Sugahara, M, Nakane, T, Yamashita, K, Hirata, K, Ueno, G, Kimura, T, Hisano, T, Muramoto, K, Sawai, H, Takeda, H, Mizohata, E, Yamashita, A, Kanematsu, Y, Takano, Y, Nango, E, Tanaka, R, Nureki, O, Ikemoto, Y, Murakami, H, Owada, S, Tono, K, Yabashi, M, Yamamoto, M, Ago, H, Iwata, S, Sugimoto, H, Shiro, Y, Kubo, M. | | Deposit date: | 2017-08-09 | | Release date: | 2017-12-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Capturing an initial intermediate during the P450nor enzymatic reaction using time-resolved XFEL crystallography and caged-substrate.
Nat Commun, 8, 2017
|
|
7AIP
 
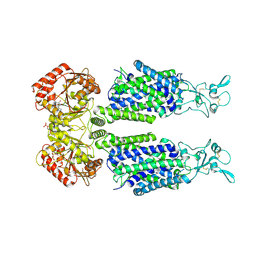 | | Structure of Human Potassium Chloride Transporter KCC1 in NaCl (Reference Map) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Ebenhoch, R, Chi, G, Man, H, Wang, D, McKinley, G, Mukhopadhyay, S.M.M, MacLean, E.M, Chalk, R, Moreau, C, Snee, M, Bohstedt, T, Liko, I, Tehan, B.G, Almeida, F.G, Elkins, J, Singh, N.K, Abrusci, P, Arrowsmith, C.H, Tang, H, Robinson, C.V, Bountra, C, Edwards, A.M, Marsden, B.D, Burgess-Brown, N.A, Duerr, K.L, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-09-28 | | Release date: | 2021-06-02 | | Last modified: | 2021-07-28 | | Method: | ELECTRON MICROSCOPY (3.12 Å) | | Cite: | Phospho-regulation, nucleotide binding and ion access control in potassium-chloride cotransporters.
Embo J., 40, 2021
|
|
6PSS
 
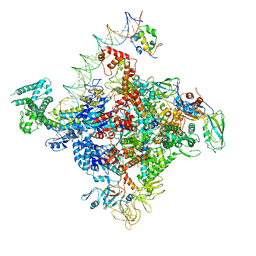 | | Escherichia coli RNA polymerase promoter unwinding intermediate (TRPi1.5a) with TraR and mutant rpsT P2 promoter | | Descriptor: | DNA (85-MER), DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Chen, J, Chiu, C.E, Campbell, E.A, Darst, S.A. | | Deposit date: | 2019-07-13 | | Release date: | 2020-03-25 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Stepwise Promoter Melting by Bacterial RNA Polymerase.
Mol.Cell, 78, 2020
|
|
6ET9
 
 | | Structure of the acetoacetyl-CoA-thiolase/HMG-CoA-synthase complex from Methanothermococcus thermolithotrophicus at 2.75 A | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Acetyl-CoA acetyltransferase thiolase, CHLORIDE ION, ... | | Authors: | Engilberge, S, Voegeli, B, Girard, E, Riobe, F, Maury, O, Erb, T.J, Shima, S, Wagner, T. | | Deposit date: | 2017-10-25 | | Release date: | 2018-03-14 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Archaeal acetoacetyl-CoA thiolase/HMG-CoA synthase complex channels the intermediate via a fused CoA-binding site.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
2INS
 
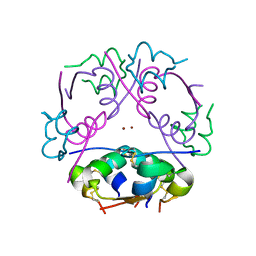 | | THE STRUCTURE OF DES-PHE B1 BOVINE INSULIN | | Descriptor: | DES-PHE B1 INSULIN (CHAIN A), DES-PHE B1 INSULIN (CHAIN B), ZINC ION | | Authors: | Smith, G.D, Duax, W.L, Dodson, E.J, Dodson, G.G, Degraaf, R.A.G, Reynolds, C.D. | | Deposit date: | 1982-05-10 | | Release date: | 1982-08-05 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The Structure of Des-Phe B1 Bovine Insulin
Acta Crystallogr.,Sect.B, 38, 1982
|
|
4S39
 
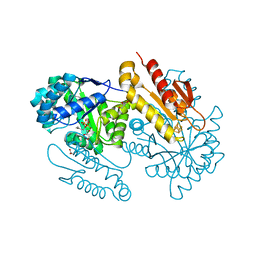 | | IspG in complex with HMBPP | | Descriptor: | (2E)-4-hydroxy-3-methylbut-2-en-1-yl trihydrogen diphosphate, 4-hydroxy-3-methylbut-2-en-1-yl diphosphate synthase, GLYCEROL, ... | | Authors: | Quitterer, F, Frank, A, Wang, K, Guodong, R, O'Dowd, B, Li, J, Guerra, F, Abdel-Azeim, S, Bacher, A, Eppinger, J, Oldfield, E, Groll, M. | | Deposit date: | 2015-01-26 | | Release date: | 2015-04-29 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Atomic-Resolution Structures of Discrete Stages on the Reaction Coordinate of the [Fe4S4] Enzyme IspG (GcpE).
J.Mol.Biol., 427, 2015
|
|
5Y7H
 
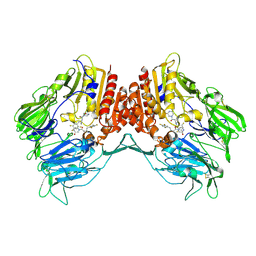 | | Crystal structure of human DPP4 in complex with inhibitor3 | | Descriptor: | (R)-4-(3-amino-4-(2,4,5-trifluorophenyl)butanoyl)piperazin-2-one, Dipeptidyl peptidase 4 | | Authors: | Lee, H.K, Kim, E.E. | | Deposit date: | 2017-08-17 | | Release date: | 2019-03-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Unique binding mode of Evogliptin with human dipeptidyl peptidase IV.
Biochem.Biophys.Res.Commun., 494, 2017
|
|
6B86
 
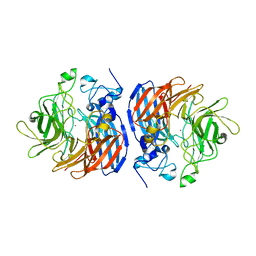 | | 2.2A Crystal Structure of Co-CAO1 | | Descriptor: | COBALT (II) ION, Carotenoid oxygenase 1 | | Authors: | Hill, H.E, Kiser, P.D. | | Deposit date: | 2017-10-05 | | Release date: | 2018-07-04 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Preparation and characterization of metal-substituted carotenoid cleavage oxygenases.
J. Biol. Inorg. Chem., 23, 2018
|
|
5N0H
 
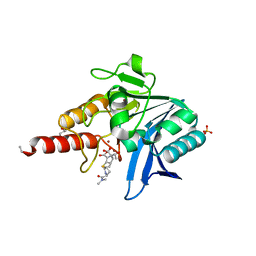 | | Crystal structure of NDM-1 in complex with hydrolyzed meropenem - new refinement | | Descriptor: | (2S,3R)-2-[(2S,3R)-1,3-bis(oxidanyl)-1-oxidanylidene-butan-2-yl]-4-[(3S,5S)-5-(dimethylcarbamoyl)pyrrolidin-3-yl]sulfan yl-3-methyl-2,3-dihydro-1H-pyrrole-5-carboxylic acid, GLYCEROL, Metallo-beta-lactamase type 2, ... | | Authors: | Raczynska, J.E, Shabalin, I.G, Jaskolski, M, Minor, W, Wlodawer, A, King, D.T, Strynadka, N.C.J. | | Deposit date: | 2017-02-03 | | Release date: | 2017-04-05 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A close look onto structural models and primary ligands of metallo-beta-lactamases.
Drug Resist. Updat., 40, 2018
|
|
7AY6
 
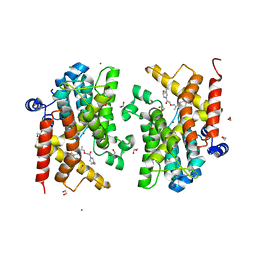 | | Crystal structure of the PDE4D catalytic domain in complex with GEBR-41b | | Descriptor: | 1,2-ETHANEDIOL, 2-[(~{Z})-1-(3-cyclopentyloxy-4-methoxy-phenyl)ethylideneamino]oxy-1-[(2~{S},6~{S})-2,6-dimethylmorpholin-4-yl]ethanone, MAGNESIUM ION, ... | | Authors: | Torretta, A, Abbate, S, Parisini, E. | | Deposit date: | 2020-11-11 | | Release date: | 2021-06-30 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Design, synthesis, biological evaluation and structural characterization of novel GEBR library PDE4D inhibitors.
Eur.J.Med.Chem., 223, 2021
|
|
4TN8
 
 | | Crystal structure of Thermus Thermophilus thioredoxin solved by sulfur SAD using Swiss Light Source data | | Descriptor: | CHLORIDE ION, Thioredoxin | | Authors: | Weinert, T, Waltersperger, S, Olieric, V, Panepucci, E, Chen, L, Rose, J.P, Wang, M, Wang, B.C, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2014-06-03 | | Release date: | 2014-12-10 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Fast native-SAD phasing for routine macromolecular structure determination.
Nat.Methods, 12, 2015
|
|
6PSQ
 
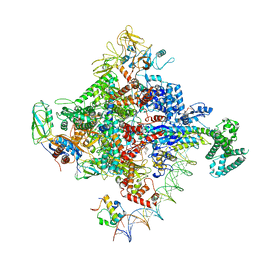 | | Escherichia coli RNA polymerase closed complex (TRPc) with TraR and rpsT P2 promoter | | Descriptor: | CHAPSO, DNA (85-MER), DNA-directed RNA polymerase subunit alpha, ... | | Authors: | Chen, J, Chiu, C.E, Campbell, E.A, Darst, S.A. | | Deposit date: | 2019-07-13 | | Release date: | 2020-03-25 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Stepwise Promoter Melting by Bacterial RNA Polymerase.
Mol.Cell, 78, 2020
|
|
4GC4
 
 | | Crystal structure of the mutant R160A.R203A of orotidine 5'-monophosphate decarboxylase from Methanobacterium thermoautotrophicum complexed with inhibitor BMP | | Descriptor: | 6-HYDROXYURIDINE-5'-PHOSPHATE, Orotidine 5'-phosphate decarboxylase | | Authors: | Fedorov, A.A, Fedorov, E.V, Desai, B, Gerlt, J.A, Almo, S.C. | | Deposit date: | 2012-07-29 | | Release date: | 2013-06-19 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.4204 Å) | | Cite: | Conformational changes in orotidine 5'-monophosphate decarboxylase: a structure-based explanation for how the 5'-phosphate group activates the enzyme.
Biochemistry, 51, 2012
|
|
4TQ6
 
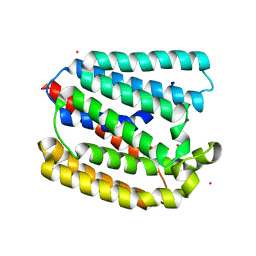 | | Structure of a UbiA homolog from Archaeoglobus fulgidus bound to Cd2+ | | Descriptor: | CADMIUM ION, prenyltransferase | | Authors: | Huang, H, Levin, E.J, Bai, Y, Zhou, M, New York Consortium on Membrane Protein Structure (NYCOMPS) | | Deposit date: | 2014-06-10 | | Release date: | 2014-07-16 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.0678 Å) | | Cite: | Structure of a Membrane-Embedded Prenyltransferase Homologous to UBIAD1.
Plos Biol., 12, 2014
|
|
