6VFX
 
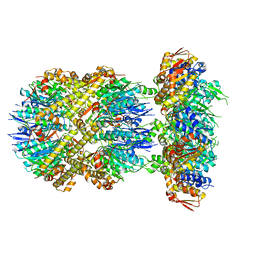 | | ClpXP from Neisseria meningitidis - Conformation B | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP-dependent Clp protease ATP-binding subunit ClpX, ... | | 著者 | Ripstein, Z.A, Vahidi, S, Houry, W.A, Rubinstein, J.L, Kay, L.E. | | 登録日 | 2020-01-06 | | 公開日 | 2020-01-22 | | 最終更新日 | 2024-03-06 | | 実験手法 | ELECTRON MICROSCOPY (2.9 Å) | | 主引用文献 | A processive rotary mechanism couples substrate unfolding and proteolysis in the ClpXP degradation machinery.
Elife, 9, 2020
|
|
6VHQ
 
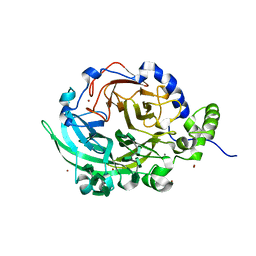 | | Crystal structure of Bacillus subtilis levansucrase (D86A/E342A) in complex with oligosaccharides | | 分子名称: | BROMIDE ION, CALCIUM ION, Glycoside hydrolase family 68 protein, ... | | 著者 | Diaz-Vilchis, A, Raga-Carbajal, E, Rojas-Trejo, S, Olvera, C, Rudino-Pinera, E. | | 登録日 | 2020-01-10 | | 公開日 | 2021-01-13 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2.047 Å) | | 主引用文献 | The molecular basis of the nonprocessive elongation mechanism in levansucrases.
J.Biol.Chem., 296, 2020
|
|
8ESI
 
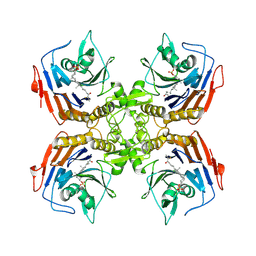 | | Bile Salt Hydrolase from B. longum with covalent inhibitor bound | | 分子名称: | (1R,3aS,3bR,5aR,7R,9aS,9bS,11aR)-1-[(2R)-6-fluoro-5-oxohexan-2-yl]-9a,11a-dimethylhexadecahydro-1H-cyclopenta[a]phenanthren-7-yl hydrogen sulfate (non-preferred name), Conjugated bile acid hydrolase | | 著者 | Walker, M.E, Lim, L, Redinbo, M.R. | | 登録日 | 2022-10-14 | | 公開日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.35 Å) | | 主引用文献 | Structural diversity of bile salt hydrolases reveals rationale for substrate selectivity
To Be Published
|
|
4Y8K
 
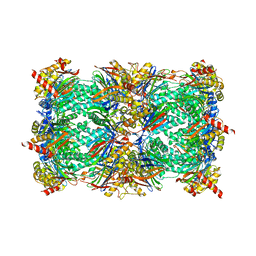 | | Yeast 20S proteasome in complex with H-APLL-ep | | 分子名称: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CHLORIDE ION, H-APLL-ep, ... | | 著者 | Huber, E.M, Groll, M. | | 登録日 | 2015-02-16 | | 公開日 | 2015-06-17 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Systematic Analyses of Substrate Preferences of 20S Proteasomes Using Peptidic Epoxyketone Inhibitors.
J.Am.Chem.Soc., 137, 2015
|
|
5JQX
 
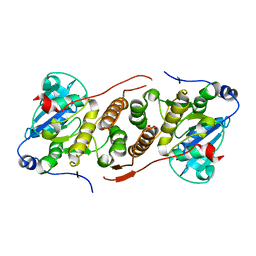 | | Crystal structure of glucosyl-3-phosphoglycerate synthase from Mycobacterium tuberculosis in complex with phosphoglyceric acid (PGA) - GpgS*PGA | | 分子名称: | 3-PHOSPHOGLYCERIC ACID, Glucosyl-3-phosphoglycerate synthase | | 著者 | Albesa-Jove, D, Sancho-Vaello, E, Rodrigo-Unzueta, A, Comino, N, Carreras-Gonzalez, A, Arrasate, P, Urresti, S, Guerin, M.E. | | 登録日 | 2016-05-05 | | 公開日 | 2017-05-24 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2.82 Å) | | 主引用文献 | Structural Snapshots and Loop Dynamics along the Catalytic Cycle of Glycosyltransferase GpgS.
Structure, 25, 2017
|
|
8HFD
 
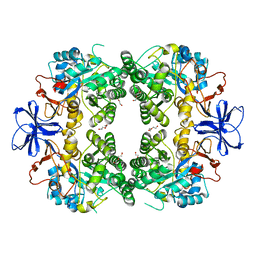 | | Crystal structure of allantoinase from E. coli BL21 | | 分子名称: | Allantoinase, DI(HYDROXYETHYL)ETHER, ZINC ION | | 著者 | Lin, E.S, Huang, H.Y, Yang, P.C, Liu, H.W, Huang, C.Y. | | 登録日 | 2022-11-10 | | 公開日 | 2023-10-18 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (2.07 Å) | | 主引用文献 | Crystal Structure of Allantoinase from Escherichia coli BL21: A Molecular Insight into a Role of the Active Site Loops in Catalysis.
Molecules, 28, 2023
|
|
3IAR
 
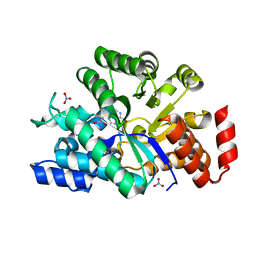 | | The crystal structure of human adenosine deaminase | | 分子名称: | (2R,3S,5R)-5-(6-amino-9H-purin-9-yl)-tetrahydro-2-(hydroxymethyl)furan-3-ol, Adenosine deaminase, GLYCEROL, ... | | 著者 | Ugochukwu, E, Zhang, Y, Hapka, E, Yue, W.W, Bray, J.E, Muniz, J, Burgess-Brown, N, Chaikuad, A, von Delft, F, Bountra, C, Arrowsmith, C.H, Weigelt, J, Edwards, A, Kavanagh, K.L, Oppermann, U, Structural Genomics Consortium (SGC) | | 登録日 | 2009-07-14 | | 公開日 | 2009-08-11 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (1.52 Å) | | 主引用文献 | The crystal structure of human adenosine deaminase
To be Published
|
|
8ETF
 
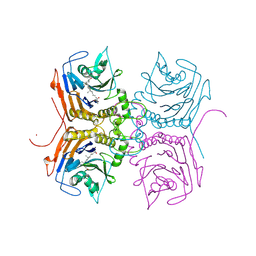 | | Bile Salt Hydrolase B from Lactobacillus gasseri with covalent inhibitor bound | | 分子名称: | (5R)-1-fluoro-5-[(1R,3aS,3bR,5aR,7R,9aS,9bS,11aR)-7-hydroxy-9a,11a-dimethylhexadecahydro-1H-cyclopenta[a]phenanthren-1-yl]hexan-2-one (non-preferred name), Choloylglycine hydrolase, NICKEL (II) ION | | 著者 | Walker, M.E, Redinbo, M.R. | | 登録日 | 2022-10-17 | | 公開日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (1.79 Å) | | 主引用文献 | Structural diversity of bile salt hydrolases reveals rationale for substrate selectivity
To Be Published
|
|
8EYG
 
 | |
8F2N
 
 | | Phi-29 partially-expanded fiberless prohead | | 分子名称: | Major capsid protein | | 著者 | Woodson, M.E, Morais, M.C, Scott, S.D, Choi, K.H, Jardine, P.J, Zhang, W. | | 登録日 | 2022-11-08 | | 公開日 | 2023-11-15 | | 実験手法 | ELECTRON MICROSCOPY (3 Å) | | 主引用文献 | Phi-29 partially-expanded fiberless prohead
To Be Published
|
|
6V8N
 
 | |
6SP2
 
 | | CryoEM structure of SERINC from Drosophila melanogaster | | 分子名称: | CARDIOLIPIN, Lauryl Maltose Neopentyl Glycol, Membrane protein TMS1d, ... | | 著者 | Pye, V.E, Nans, A, Cherepanov, P. | | 登録日 | 2019-08-30 | | 公開日 | 2020-01-01 | | 最終更新日 | 2022-03-30 | | 実験手法 | ELECTRON MICROSCOPY (3.33 Å) | | 主引用文献 | A bipartite structural organization defines the SERINC family of HIV-1 restriction factors.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6VAW
 
 | | Peanut lectin complexed with N-beta-D-galactopyranosyl-L-succinamoyl derivative (NGS) | | 分子名称: | CALCIUM ION, Galactose-binding lectin, MANGANESE (II) ION, ... | | 著者 | Otero, L.H, Primo, E.D, Cagnoni, A.J, Klinke, S, Goldbaum, F.A, Uhrig, M.L. | | 登録日 | 2019-12-18 | | 公開日 | 2020-10-28 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | Crystal structures of peanut lectin in the presence of synthetic beta-N- and beta-S-galactosides disclose evidence for the recognition of different glycomimetic ligands.
Acta Crystallogr D Struct Biol, 76, 2020
|
|
5DKN
 
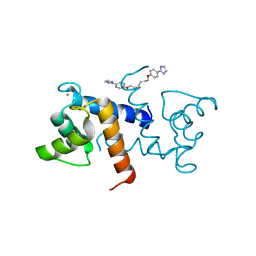 | | Crystal Structure of Calcium-loaded S100B bound to SBi4225 | | 分子名称: | 2,2'-[heptane-1,7-diylbis(oxybenzene-4,1-diyl)]bis(1H-imidazole), CALCIUM ION, Protein S100-B | | 著者 | Cavalier, M.C, Ansari, M.I, Pierce, A.D, Wilder, P.T, McKnight, L.E, Raman, E.P, Neau, D.B, Bezawada, P, Alasady, M.J, Varney, K.M, Toth, E.A, MacKerell Jr, A.D, Coop, A, Weber, D.J. | | 登録日 | 2015-09-03 | | 公開日 | 2016-01-20 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.528 Å) | | 主引用文献 | Small Molecule Inhibitors of Ca(2+)-S100B Reveal Two Protein Conformations.
J.Med.Chem., 59, 2016
|
|
5J3Q
 
 | | Crystal structure of S. pombe Dcp1:Edc1 mRNA decapping complex | | 分子名称: | Edc1, mRNA-decapping enzyme subunit 1 | | 著者 | Valkov, E, Muthukumar, S, Chang, C.T, Jonas, S, Weichenrieder, O, Izaurralde, E. | | 登録日 | 2016-03-31 | | 公開日 | 2016-05-18 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (1.87 Å) | | 主引用文献 | Structure of the Dcp2-Dcp1 mRNA-decapping complex in the activated conformation.
Nat.Struct.Mol.Biol., 23, 2016
|
|
6SP1
 
 | | KEAP1 IN COMPLEX WITH COMPOUND 6 | | 分子名称: | (1~{S},2~{R})-2-[[(1~{S})-1-[[1,3-bis(oxidanylidene)isoindol-2-yl]methyl]-5-(2-hydroxyethyloxy)-3,4-dihydro-1~{H}-isoquinolin-2-yl]carbonyl]cyclohexane-1-carboxylic acid, ACETATE ION, Kelch-like ECH-associated protein 1 | | 著者 | Ontoria, J.M, Biancofiore, I, Fezzardi, P, Torrente de Haro, E, Colarusso, S, Bianchi, E, Andreini, M, Patsilinakos, A, Summa, V, Pacifici, R, Munoz-Sanjuan, I, Park, L, Bresciani, A, Dominguez, C, Toledo-Sherman, L, Harper, S. | | 登録日 | 2019-08-30 | | 公開日 | 2020-06-03 | | 実験手法 | X-RAY DIFFRACTION (2.57 Å) | | 主引用文献 | Combined Peptide and Small-Molecule Approach toward Nonacidic THIQ Inhibitors of the KEAP1/NRF2 Interaction.
Acs Med.Chem.Lett., 11, 2020
|
|
6VFS
 
 | | ClpXP from Neisseria meningitidis - Conformation A | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP-dependent Clp protease ATP-binding subunit ClpX, ... | | 著者 | Ripstein, Z.A, Vahidi, S, Houry, W.A, Rubinstein, J.L, Kay, L.E. | | 登録日 | 2020-01-06 | | 公開日 | 2020-01-22 | | 最終更新日 | 2024-03-06 | | 実験手法 | ELECTRON MICROSCOPY (3.3 Å) | | 主引用文献 | A processive rotary mechanism couples substrate unfolding and proteolysis in the ClpXP degradation machinery.
Elife, 9, 2020
|
|
6T0L
 
 | | Crystal structure of CYP124 in complex with inhibitor compound 5' | | 分子名称: | CHLORIDE ION, CYP124 in complex with inhibitor compound 5', DI(HYDROXYETHYL)ETHER, ... | | 著者 | Bukhdruker, S, Marin, E, Varaksa, T, Gilep, A, Strushkevich, N, Borshchevskiy, V. | | 登録日 | 2019-10-03 | | 公開日 | 2020-10-14 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Metabolic Fate of Human Immunoactive Sterols in Mycobacterium tuberculosis.
J.Mol.Biol., 433, 2021
|
|
6VHL
 
 | | Paired Helical Filament from Alzheimer's Disease Human Brain Tissue | | 分子名称: | GLYCINE, Microtubule-associated protein tau | | 著者 | Arakhamia, T, Lee, C.E, Carlomagno, Y, Duong, D.M, Kundinger, S.R, Wang, K, Williams, D, DeTure, M, Dickson, D.W, Cook, C.N, Seyfried, N.T, Petrucelli, L, Fitzpatrick, A.W.P. | | 登録日 | 2020-01-10 | | 公開日 | 2020-03-04 | | 最終更新日 | 2024-03-06 | | 実験手法 | ELECTRON MICROSCOPY (3.3 Å) | | 主引用文献 | Posttranslational Modifications Mediate the Structural Diversity of Tauopathy Strains.
Cell, 180, 2020
|
|
6PLR
 
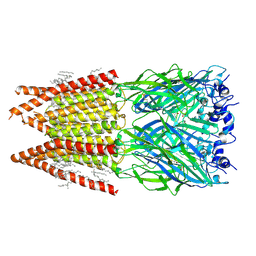 | | CryoEM structure of zebra fish alpha-1 glycine receptor bound with glycine in nanodisc, desensitized state | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCINE, Glycine receptor subunit alphaZ1, ... | | 著者 | Yu, J, Zhu, H, Gouaux, E. | | 登録日 | 2019-07-01 | | 公開日 | 2021-01-06 | | 最終更新日 | 2021-07-21 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | Mechanism of gating and partial agonist action in the glycine receptor.
Cell, 184, 2021
|
|
5N1Q
 
 | | METHYL-COENZYME M REDUCTASE III FROM METHANOTHERMOCOCCUS THERMOLITHOTROPHICUS AT 1.9 A RESOLUTION | | 分子名称: | 1-THIOETHANESULFONIC ACID, Coenzyme B, FACTOR 430, ... | | 著者 | Wagner, T, Wegner, C.E, Ermler, U, Shima, S. | | 登録日 | 2017-02-06 | | 公開日 | 2017-06-14 | | 最終更新日 | 2024-05-01 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Phylogenetic and Structural Comparisons of the Three Types of Methyl Coenzyme M Reductase from Methanococcales and Methanobacteriales.
J.Bacteriol., 199, 2017
|
|
7ULA
 
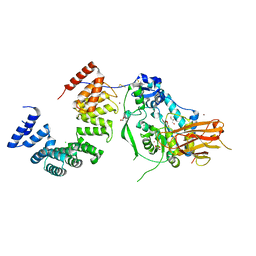 | | Structure of the Pseudomonas putida AlgKX modification and secretion complex | | 分子名称: | Alginate biosynthesis protein AlgK, Alginate biosynthesis protein AlgX, CHLORIDE ION, ... | | 著者 | Gheorghita, A.A, Li, E.Y, Pfoh, R, Howell, P.L. | | 登録日 | 2022-04-04 | | 公開日 | 2022-12-14 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.46 Å) | | 主引用文献 | Structure of the AlgKX modification and secretion complex required for alginate production and biofilm attachment in Pseudomonas aeruginosa.
Nat Commun, 13, 2022
|
|
6PS1
 
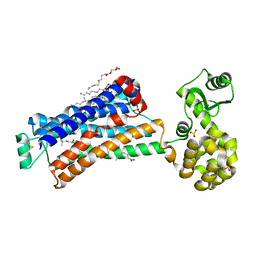 | | XFEL beta2 AR structure by ligand exchange from Alprenolol to Timolol. | | 分子名称: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (2S)-1-(tert-butylamino)-3-[(4-morpholin-4-yl-1,2,5-thiadiazol-3-yl)oxy]propan-2-ol, CHOLESTEROL, ... | | 著者 | Ishchenko, A, Stauch, B, Han, G.W, Batyuk, A, Shiriaeva, A, Li, C, Zatsepin, N.A, Weierstall, U, Liu, W, Nango, E, Nakane, T, Tanaka, R, Tono, K, Joti, Y, Iwata, S, Moraes, I, Gati, C, Cherezov, C. | | 登録日 | 2019-07-12 | | 公開日 | 2019-11-13 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (3.2 Å) | | 主引用文献 | Toward G protein-coupled receptor structure-based drug design using X-ray lasers.
Iucrj, 6, 2019
|
|
6VAU
 
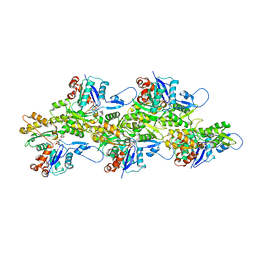 | | Bare actin filament from a partially cofilin-decorated sample | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, Actin, alpha skeletal muscle, ... | | 著者 | Huehn, A.R, Bibeau, J.P, Schramm, A.C, Cao, W, De La Cruz, E.M, Sindelar, C.V. | | 登録日 | 2019-12-17 | | 公開日 | 2020-01-01 | | 最終更新日 | 2024-03-06 | | 実験手法 | ELECTRON MICROSCOPY (3.5 Å) | | 主引用文献 | Structures of cofilin-induced structural changes reveal local and asymmetric perturbations of actin filaments.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6SNB
 
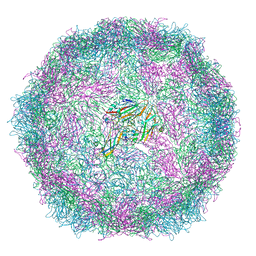 | | Structure of Coxsackievirus A10 A-particle | | 分子名称: | Capsid protein VP1, Capsid protein VP2, Capsid protein VP3 | | 著者 | Zhao, Y, Zhou, D, Ni, T, Karia, D, Kotecha, A, Wang, X, Rao, Z, Jones, E.Y, Fry, E.E, Ren, J, Stuart, D.I. | | 登録日 | 2019-08-23 | | 公開日 | 2020-01-15 | | 最終更新日 | 2024-05-22 | | 実験手法 | ELECTRON MICROSCOPY (4.4 Å) | | 主引用文献 | Hand-foot-and-mouth disease virus receptor KREMEN1 binds the canyon of Coxsackie Virus A10.
Nat Commun, 11, 2020
|
|
