5ACF
 
 | | X-ray Structure of LPMO | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, COPPER (II) ION, ... | | Authors: | Frandsen, K.E.H, Poulsen, J.N, Tovborg, M, Johansen, K.S, Lo Leggio, L. | | Deposit date: | 2015-08-17 | | Release date: | 2016-03-02 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The molecular basis of polysaccharide cleavage by lytic polysaccharide monooxygenases.
Nat. Chem. Biol., 12, 2016
|
|
7UJX
 
 | | Structure of cAMP-dependent protein kinase using a MD-MX procedure, produced using 2.4 Angstrom data | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Wych, D.C, Aoto, P.C, Wall, M.E. | | Deposit date: | 2022-03-31 | | Release date: | 2022-12-07 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Molecular-dynamics simulation methods for macromolecular crystallography.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
5NSD
 
 | | Co-crystal structure of NAMPT dimer with KPT-9274 | | Descriptor: | (~{E})-3-(6-azanylpyridin-3-yl)-~{N}-[[5-[4-[4,4-bis(fluoranyl)piperidin-1-yl]carbonylphenyl]-7-(4-fluorophenyl)-1-benzofuran-2-yl]methyl]prop-2-enamide, GLYCEROL, Nicotinamide phosphoribosyltransferase, ... | | Authors: | Neggers, J.E, Kwanten, B, Dierckx, T, Noguchi, H, Voet, A, Vercruysse, T, Baloglu, E, Senapedis, W, Jacquemyn, M, Daelemans, D. | | Deposit date: | 2017-04-26 | | Release date: | 2018-02-14 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.046 Å) | | Cite: | Target identification of small molecules using large-scale CRISPR-Cas mutagenesis scanning of essential genes.
Nat Commun, 9, 2018
|
|
5A5R
 
 | | Crystal structure of human ATAD2 bromodomain in complex with 5-5- methoxypyridin-3-yl-3-methyl-8-piperidin-4-ylamino-1,2-dihydro-1,7- naphthyridin-2-one | | Descriptor: | 1,2-ETHANEDIOL, 5-(5-methoxypyridin-3-yl)-3-methyl-8-[(piperidin-4-yl)amino]-1,2-dihydro-1,7-naphthyridin-2-one, ATPASE FAMILY AAA DOMAIN-CONTAINING PROTEIN 2, ... | | Authors: | Chung, C, Bamborough, P, Demont, E. | | Deposit date: | 2015-06-20 | | Release date: | 2015-07-22 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Fragment-Based Discovery of Low-Micromolar Atad2 Bromodomain Inhibitors.
J.Med.Chem., 58, 2015
|
|
3LHS
 
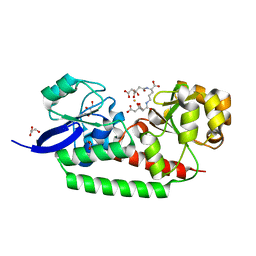 | | Open Conformation of HtsA Complexed with Staphyloferrin A | | Descriptor: | (2R)-2-(2-{[(1R)-1-carboxy-4-{[(3S)-3,4-dicarboxy-3-hydroxybutanoyl]amino}butyl]amino}-2-oxoethyl)-2-hydroxybutanedioic acid, FE (III) ION, Ferrichrome ABC transporter lipoprotein, ... | | Authors: | Grigg, J.C, Murphy, M.E.P. | | Deposit date: | 2010-01-23 | | Release date: | 2010-02-09 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | The Staphylococcus aureus siderophore receptor HtsA undergoes localized conformational changes to enclose staphyloferrin A in an arginine-rich binding pocket.
J.Biol.Chem., 285, 2010
|
|
8W88
 
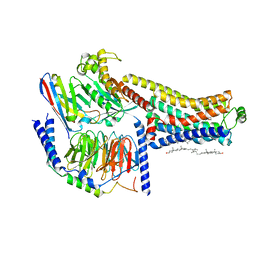 | | Cryo-EM structure of the SEP363856-bound TAAR1-Gs complex | | Descriptor: | 1-[(7~{S})-5,7-dihydro-4~{H}-thieno[2,3-c]pyran-7-yl]-~{N}-methyl-methanamine, CHOLESTEROL, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Liu, H, Zheng, Y, Wang, Y, Wang, Y, He, X, Xu, P, Huang, S, Yuan, Q, Zhang, X, Wang, S, Xu, H.E, Xu, F. | | Deposit date: | 2023-09-01 | | Release date: | 2023-11-22 | | Last modified: | 2024-01-03 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Recognition of methamphetamine and other amines by trace amine receptor TAAR1.
Nature, 624, 2023
|
|
5AJL
 
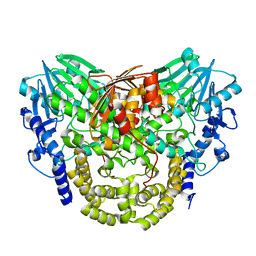 | | Sdsa sulfatase tetragonal | | Descriptor: | ALKYL SULFATASE, ZINC ION | | Authors: | De la Mora, E, Flores-Hernandez, E, Jakoncic, J, Stojanoff, V, Sanchez-Puig, N, Moreno, A. | | Deposit date: | 2015-02-25 | | Release date: | 2015-10-07 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.45 Å) | | Cite: | Sdsa Polymorph Isolation and Improvement of Their Crystal Quality Using Nonconventional Crystallization Techniques
J.Appl.Crystallogr., 48, 2015
|
|
7V0G
 
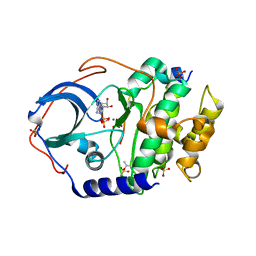 | | Structure of cAMP-dependent protein kinase using a MD-MX procedure, produced using 1.63 Angstrom data | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Wych, D.C, Aoto, P.C, Wall, M.E. | | Deposit date: | 2022-05-10 | | Release date: | 2022-12-07 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Molecular-dynamics simulation methods for macromolecular crystallography.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
8W87
 
 | | Cryo-EM structure of the METH-TAAR1 complex | | Descriptor: | (2S)-N-methyl-1-phenylpropan-2-amine, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Liu, H, Zheng, Y, Wang, Y, Wang, Y, He, X, Xu, P, Huang, S, Yuan, Q, Zhang, X, Wang, S, Xu, H.E, Xu, F. | | Deposit date: | 2023-09-01 | | Release date: | 2023-11-22 | | Last modified: | 2024-01-03 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Recognition of methamphetamine and other amines by trace amine receptor TAAR1.
Nature, 624, 2023
|
|
8W8A
 
 | | Cryo-EM structure of the RO5256390-TAAR1 complex | | Descriptor: | (4S)-4-[(2S)-2-phenylbutyl]-1,3-oxazolidin-2-imine, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Liu, H, Zheng, Y, Wang, Y, Wang, Y, He, X, Xu, P, Huang, S, Yuan, Q, Zhang, X, Wang, S, Xu, H.E, Xu, F. | | Deposit date: | 2023-09-01 | | Release date: | 2023-11-22 | | Last modified: | 2024-01-03 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Recognition of methamphetamine and other amines by trace amine receptor TAAR1.
Nature, 624, 2023
|
|
7UV0
 
 | | Structure of the sodium/iodide symporter (NIS) in complex with iodide and sodium | | Descriptor: | 1,2-DIACYL-GLYCEROL-3-SN-PHOSPHATE, IODIDE ION, SODIUM ION, ... | | Authors: | Ravera, S, Nicola, J.P, Salazar-De Simone, G, Sigworth, F, Karakas, E, Amzel, L.M, Bianchet, M, Carrasco, N. | | Deposit date: | 2022-04-29 | | Release date: | 2022-12-21 | | Last modified: | 2023-01-04 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural insights into the mechanism of the sodium/iodide symporter.
Nature, 612, 2022
|
|
6F55
 
 | | Complex structure of PACSIN SH3 domain and TRPV4 proline rich region | | Descriptor: | PACSIN 3, PRR | | Authors: | Glogowski, N.A, Goretzki, B, Diehl, E, Duchardt-Ferner, E, Hacker, C, Hellmich, U.A. | | Deposit date: | 2017-11-30 | | Release date: | 2018-10-03 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural Basis of TRPV4 N Terminus Interaction with Syndapin/PACSIN1-3 and PIP2.
Structure, 26, 2018
|
|
8UX1
 
 | |
2P85
 
 | |
1FE8
 
 | | CRYSTAL STRUCTURE OF THE VON WILLEBRAND FACTOR A3 DOMAIN IN COMPLEX WITH A FAB FRAGMENT OF IGG RU5 THAT INHIBITS COLLAGEN BINDING | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CACODYLATE ION, IMMUNOGLOBULIN IGG RU5, ... | | Authors: | Bouma, B, Huizinga, E.G, Schiphorst, M.E, Sixma, J.J, Kroon, J, Gros, P. | | Deposit date: | 2000-07-21 | | Release date: | 2001-04-04 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Identification of the collagen-binding site of the von Willebrand factor A3-domain.
J.Biol.Chem., 276, 2001
|
|
8W89
 
 | | Cryo-EM structure of the PEA-bound TAAR1-Gs complex | | Descriptor: | 2-PHENYLETHYLAMINE, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Liu, H, Zheng, Y, Wang, Y, Wang, Y, He, X, Xu, P, Huang, S, Yuan, Q, Zhang, X, Wang, S, Xu, H.E, Xu, F. | | Deposit date: | 2023-09-01 | | Release date: | 2023-11-22 | | Last modified: | 2024-01-03 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Recognition of methamphetamine and other amines by trace amine receptor TAAR1.
Nature, 624, 2023
|
|
1LLQ
 
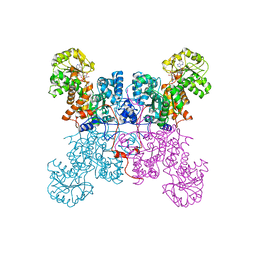 | | Crystal Structure of Malic Enzyme from Ascaris suum Complexed with Nicotinamide Adenine Dinucleotide | | Descriptor: | NAD-dependent malic enzyme, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Coleman, D.E, Jagannatha, G.S, Goldsmith, E.J, Cook, P.F, Harris, B.G. | | Deposit date: | 2002-04-29 | | Release date: | 2002-05-08 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of the malic enzyme from Ascaris suum complexed with nicotinamide adenine dinucleotide at 2.3 A resolution.
Biochemistry, 41, 2002
|
|
1DWV
 
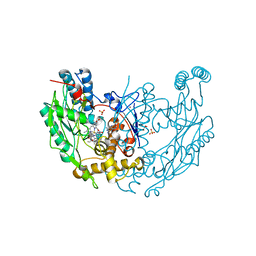 | | MURINE INDUCIBLE NITRIC OXIDE SYNTHASE OXYGENASE DIMER N-hydroxyarginine and 4-amino tetrahydrobiopterin | | Descriptor: | 2,4-DIAMINO-6-[2,3-DIHYDROXY-PROP-3-YL]-5,6,7,8-TETRAHYDROPTERIDINE, N-OMEGA-HYDROXY-L-ARGININE, NITRIC OXIDE SYNTHASE, ... | | Authors: | Crane, B.R, Arvai, A.S, Getzoff, E.D, Stuehr, D.J, Tainer, J.A. | | Deposit date: | 1999-12-14 | | Release date: | 2000-02-06 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structures of the N(Omega)-Hydroxy-L-Arginine Complex of Inducible Nitric Oxide Synthase Oxygenase Dimer with Active Andinactive Pterins
Biochemistry, 39, 2000
|
|
1FPZ
 
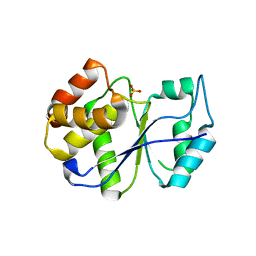 | | CRYSTAL STRUCTURE ANALYSIS OF KINASE ASSOCIATED PHOSPHATASE (KAP) WITH A SUBSTITUTION OF THE CATALYTIC SITE CYSTEINE (CYS140) TO A SERINE | | Descriptor: | CYCLIN-DEPENDENT KINASE INHIBITOR 3, SULFATE ION | | Authors: | Song, H, Hanlon, N, Brown, N.R, Noble, M.E.M, Johnson, L.N, Barford, D. | | Deposit date: | 2000-09-01 | | Release date: | 2001-05-09 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Phosphoprotein-protein interactions revealed by the crystal structure of kinase-associated phosphatase in complex with phosphoCDK2.
Mol.Cell, 7, 2001
|
|
1FHW
 
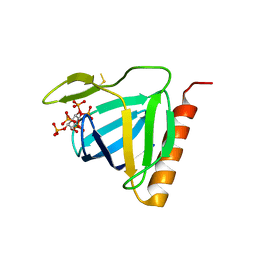 | | Structure of the pleckstrin homology domain from GRP1 in complex with inositol(1,3,4,5,6)pentakisphosphate | | Descriptor: | GUANINE NUCLEOTIDE EXCHANGE FACTOR AND INTEGRIN BINDING PROTEIN HOMOLOG GRP1, INOSITOL-(1,3,4,5,6)-PENTAKISPHOSPHATE, SULFATE ION | | Authors: | Ferguson, K.M, Kavran, J.M, Sankaran, V.G, Fournier, E, Isakoff, S.J, Skolnik, E.Y, Lemmon, M.A. | | Deposit date: | 2000-08-02 | | Release date: | 2000-08-23 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis for discrimination of 3-phosphoinositides by pleckstrin homology domains
Mol.Cell, 6, 2000
|
|
4RI4
 
 | |
4RI5
 
 | |
8PVM
 
 | | formaldehyde-inhibited [FeFe]-hydrogenase CpI from Clostridium pasteurianum, variant C299D | | Descriptor: | CHLORIDE ION, FE2/S2 (INORGANIC) CLUSTER, FORMYL GROUP, ... | | Authors: | Duan, J, Hofmann, E, Happe, T. | | Deposit date: | 2023-07-18 | | Release date: | 2023-11-29 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | Insights into the Molecular Mechanism of Formaldehyde Inhibition of [FeFe]-Hydrogenases.
J.Am.Chem.Soc., 145, 2023
|
|
1FN4
 
 | | CRYSTAL STRUCTURE OF FAB198, AN EFFICIENT PROTECTOR OF ACETYLCHOLINE RECEPTOR AGAINST MYASTHENOGENIC ANTIBODIES | | Descriptor: | MONOCLONAL ANTIBODY AGAINST ACETYLCHOLINE RECEPTOR | | Authors: | Poulas, K, Eliopoulos, E, Vatzaki, E, Navaza, J, Kontou, M. | | Deposit date: | 2000-08-21 | | Release date: | 2001-09-26 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of Fab198, an efficient protector of the acetylcholine receptor against myasthenogenic antibodies.
Eur.J.Biochem., 268, 2001
|
|
4Y80
 
 | | Yeast 20S proteasome in complex with Ac-LAI-ep | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Ac-LAI-ep, CHLORIDE ION, ... | | Authors: | Huber, E.M, Groll, M. | | Deposit date: | 2015-02-16 | | Release date: | 2015-06-17 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Systematic Analyses of Substrate Preferences of 20S Proteasomes Using Peptidic Epoxyketone Inhibitors.
J.Am.Chem.Soc., 137, 2015
|
|
